Abstract
The field of spatial transcriptomics is rapidly evolving, with increasing sample complexity, resolution, and tissue size. Yet the field lacks comprehensive solutions for automated integration and analysis of multi-slice data in either stacked (3D) or co-planar (2D) formation. To address this, we developed VR-Omics, a free, platform-agnostic software that provides end-to-end automated processing of multi-slice data through a biologist-friendly interface. Benchmarking against existing methods demonstrates VR-Omics’ unique strengths to perform comprehensive end-to-end analysis of multi-slice stacked data. Through co-planar slice analysis, VR-Omics uncovered previously undetected, dysregulated metabolic networks within rare paediatric cardiac rhabdomyomas, demonstrating its potential for biological discoveries.
Download VR-Omics for Windows 64-bit Download VR-Omics for MacOs (Beta)
Documentation
Automated Workflow
Quick Access Visualisation Features
VR-Omics Setup (Windows)
| 1 | Download VR-Omics |
Click the |
Download VR-Omics for Windows 64-bit |
|---|---|---|---|
| 2 | Download VR-Omics |
Click the |
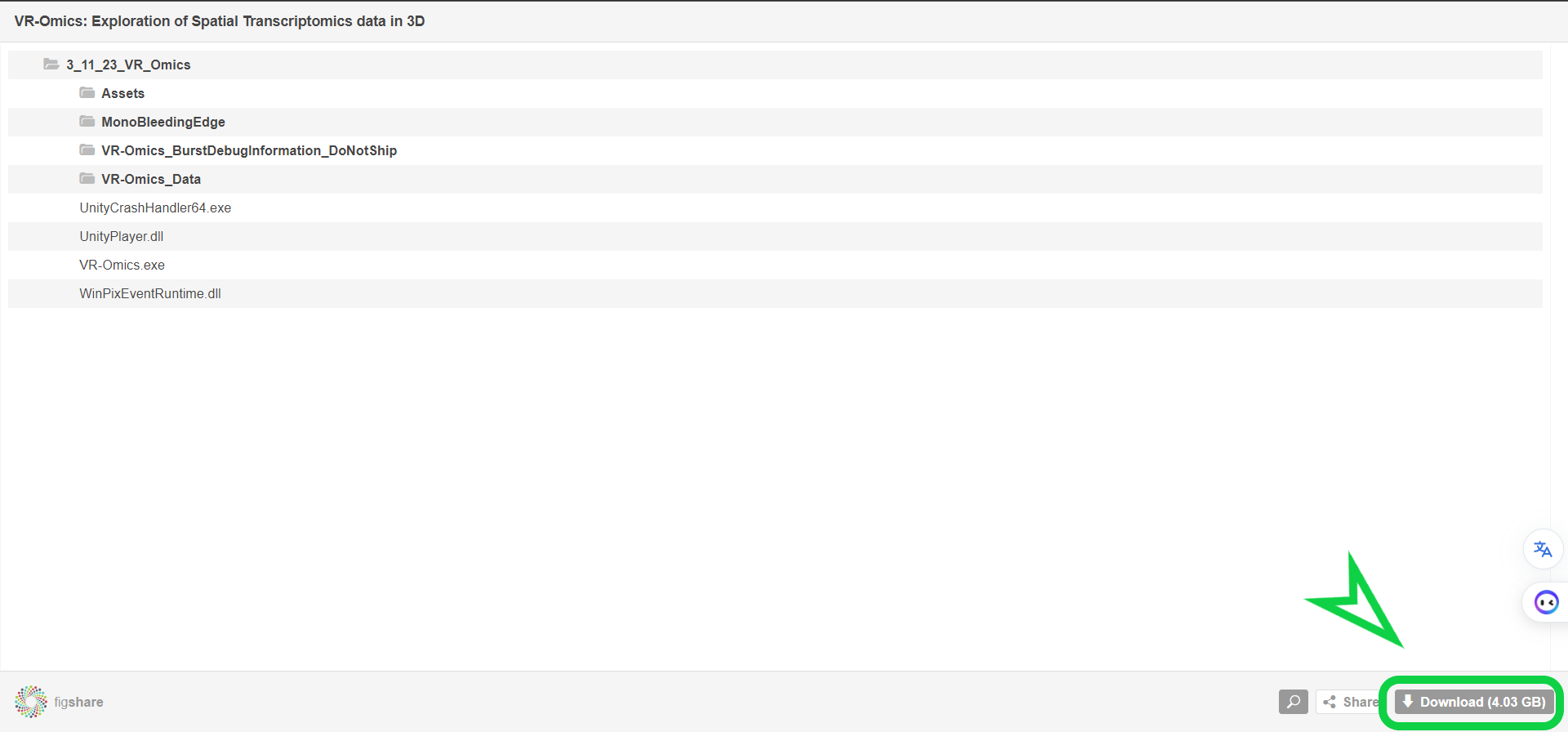
|
| 3 | Unzip the folder |
To unzip the folder, right-click on the zip file and select |
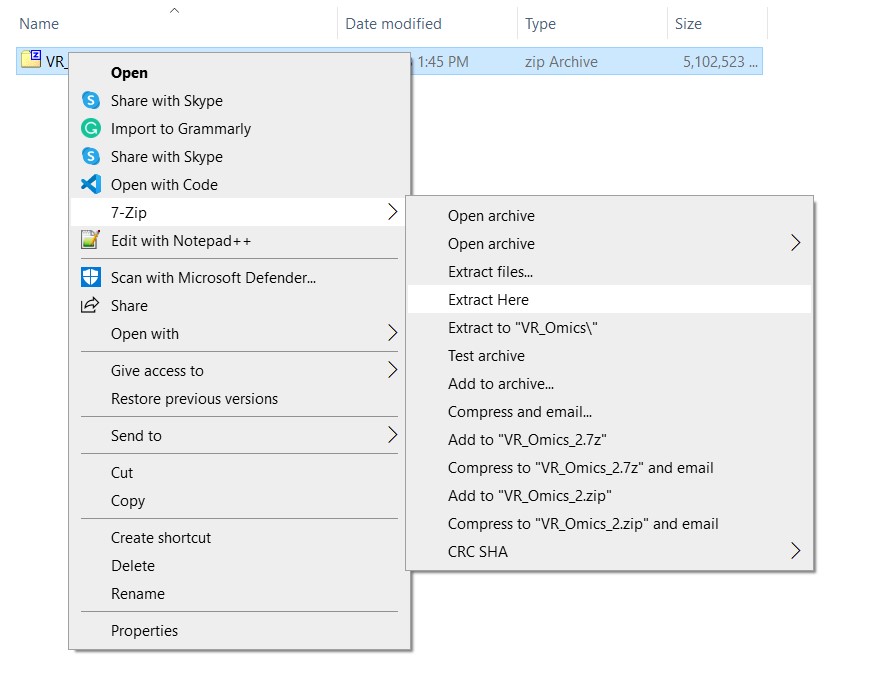
|
| 4 | Start VR-Omics |
Once the folder is unzipped, open it and double-click on the |
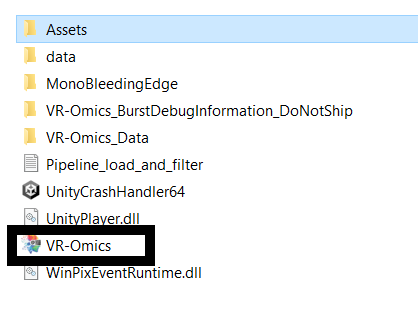
|
VR-Omics Setup (MacOs)
| 1 | Download VR-Omics |
Click the |
Download VR-Omics for MacOs (Beta) |
|---|---|---|---|
| 2 | Download VR-Omics |
Click the |
|
| 3 | Warning |
To unzip the folder, double-click on the zip file. When you try to unzip the file, you might get this warning. |
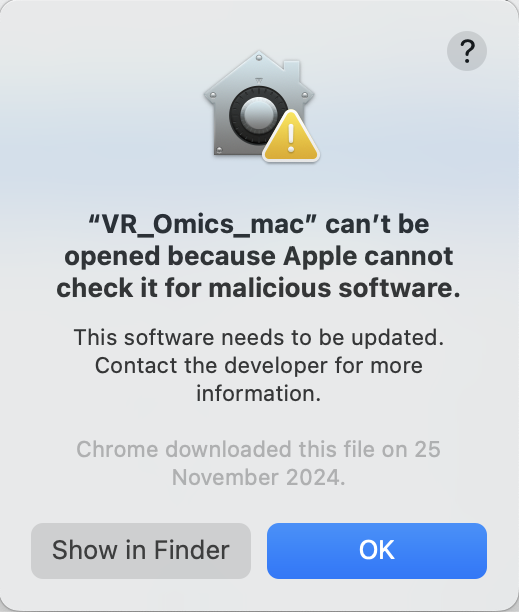
|
| 4 | Solution |
To solve the above warning, go to "Settings", "Privacy and Security", find the warning message about VR-Omics, and click |
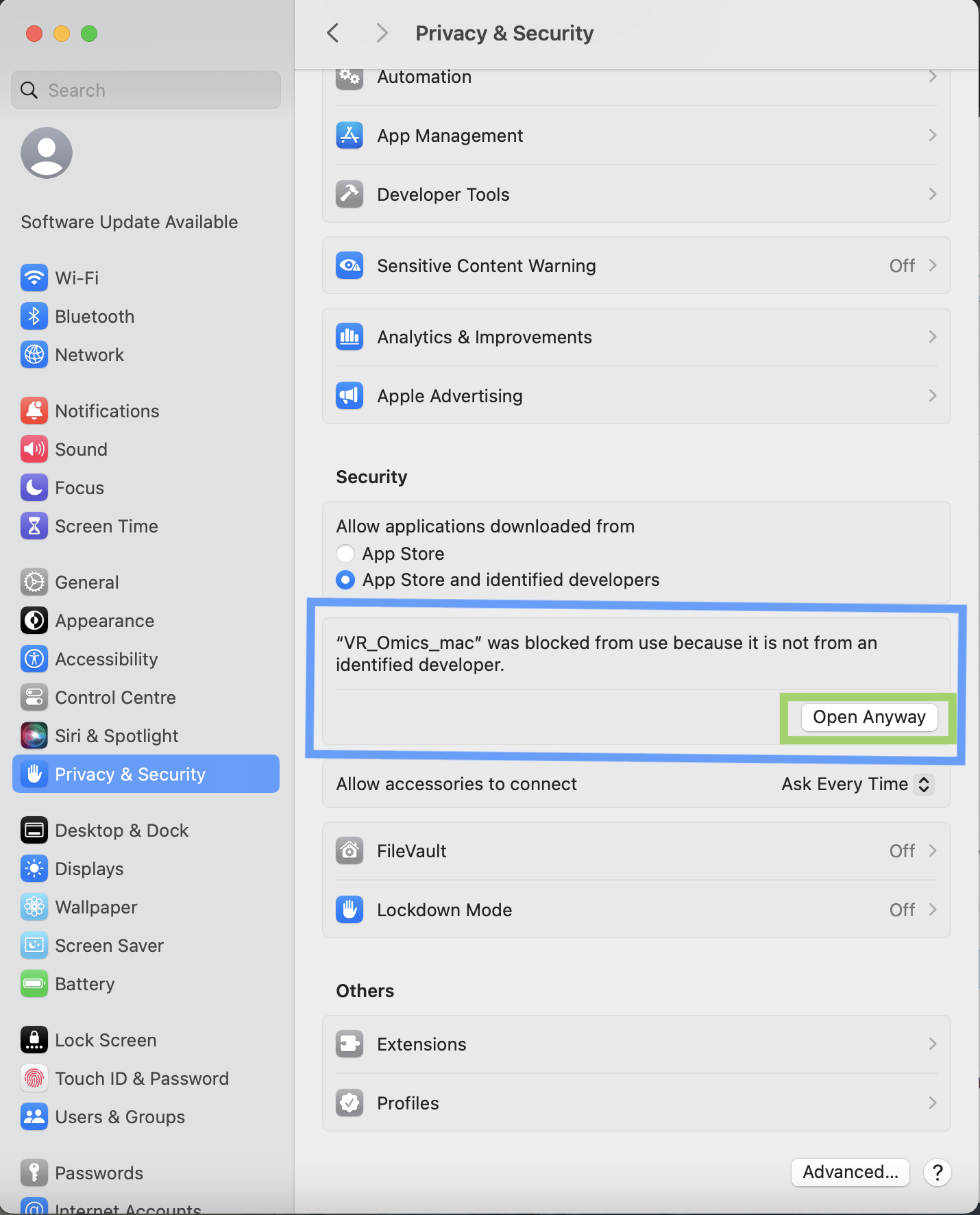
|
VR-Omics Automated Workflow
Visium
Download Data
| 1 | Choose the Download menu |
|

|
|---|---|---|---|
| 2 | Selecting datasets and filtering |
|
|
| 3 | Identify spatially variable genes |
Tick the Spatially Variable Genes box to perform spatial analysis. |
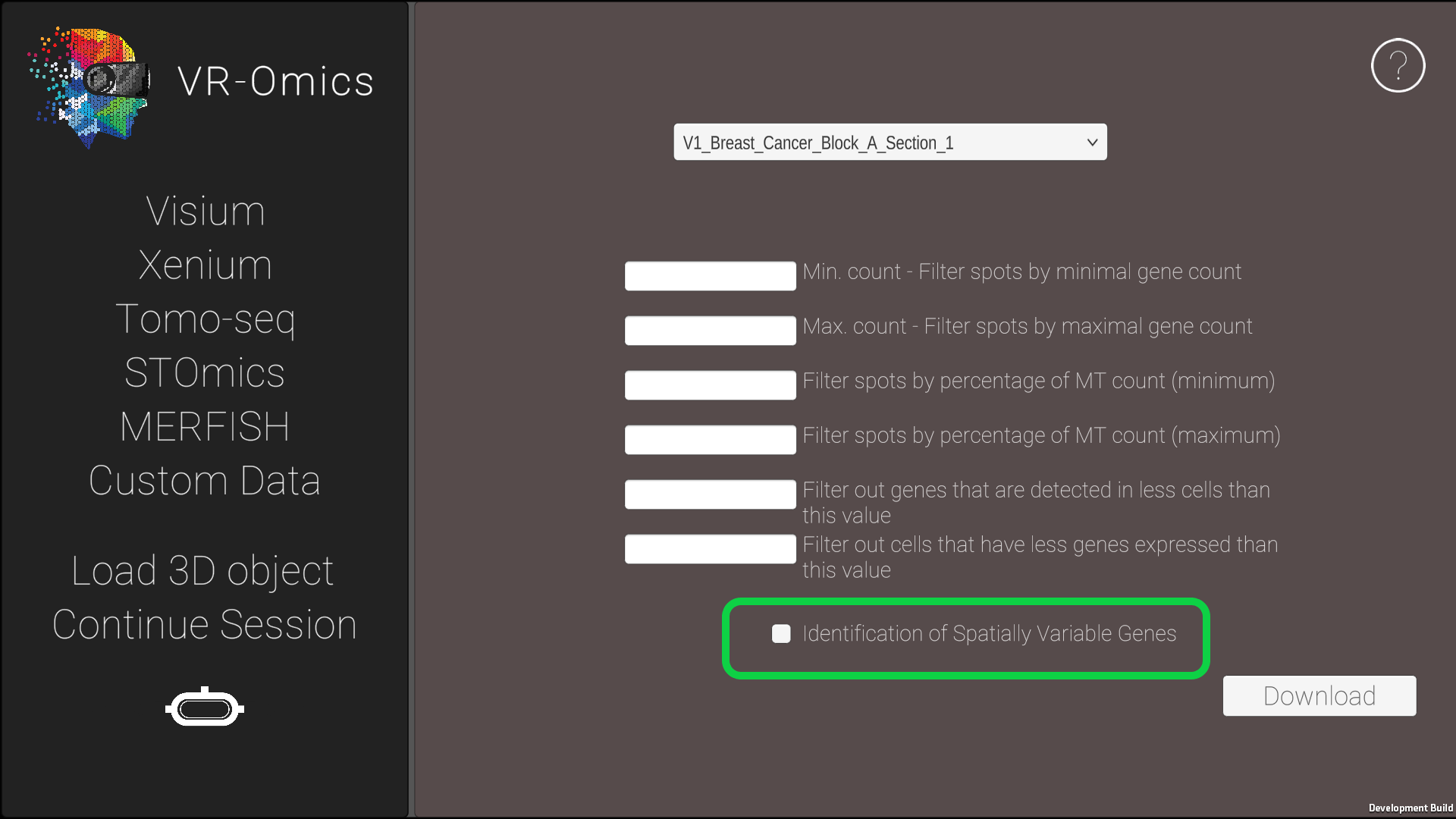
Spatially variable genes are genes that exhibit differences in expression levels across different regions or cell types within a tissue or organism. 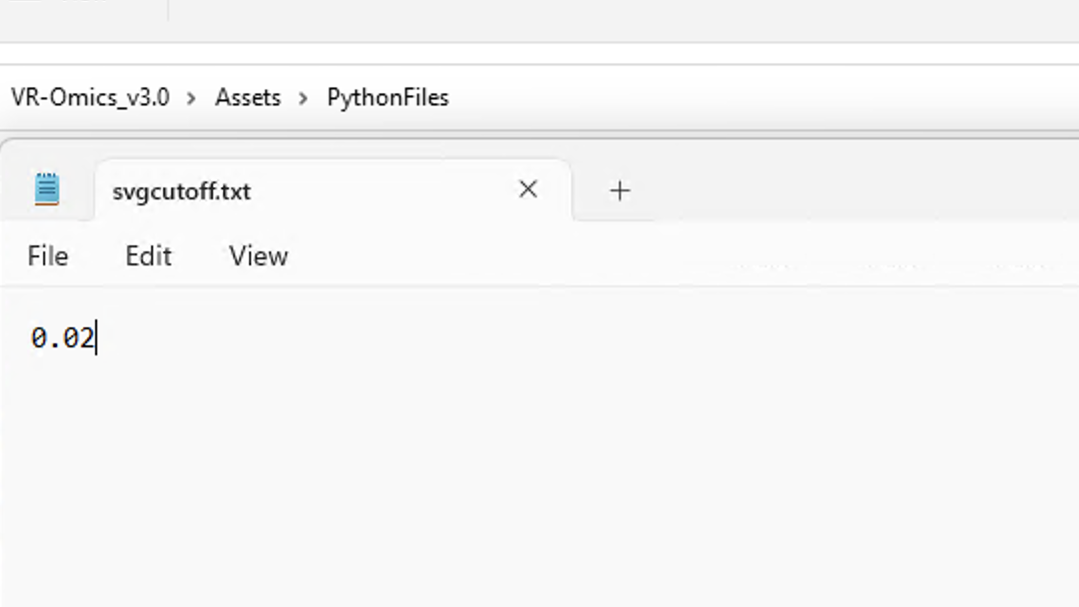
p-value threshold for SVG cutoff can be customized. To modify the p-value, go |
| 4 | Download the dataset |
Select Download to download and process the dataset. |

The data will be downloaded from the 10X Genomics Resources Website and processed to be read in VR-Omics The data will be stored in the VR-Omics folder on your local machine. 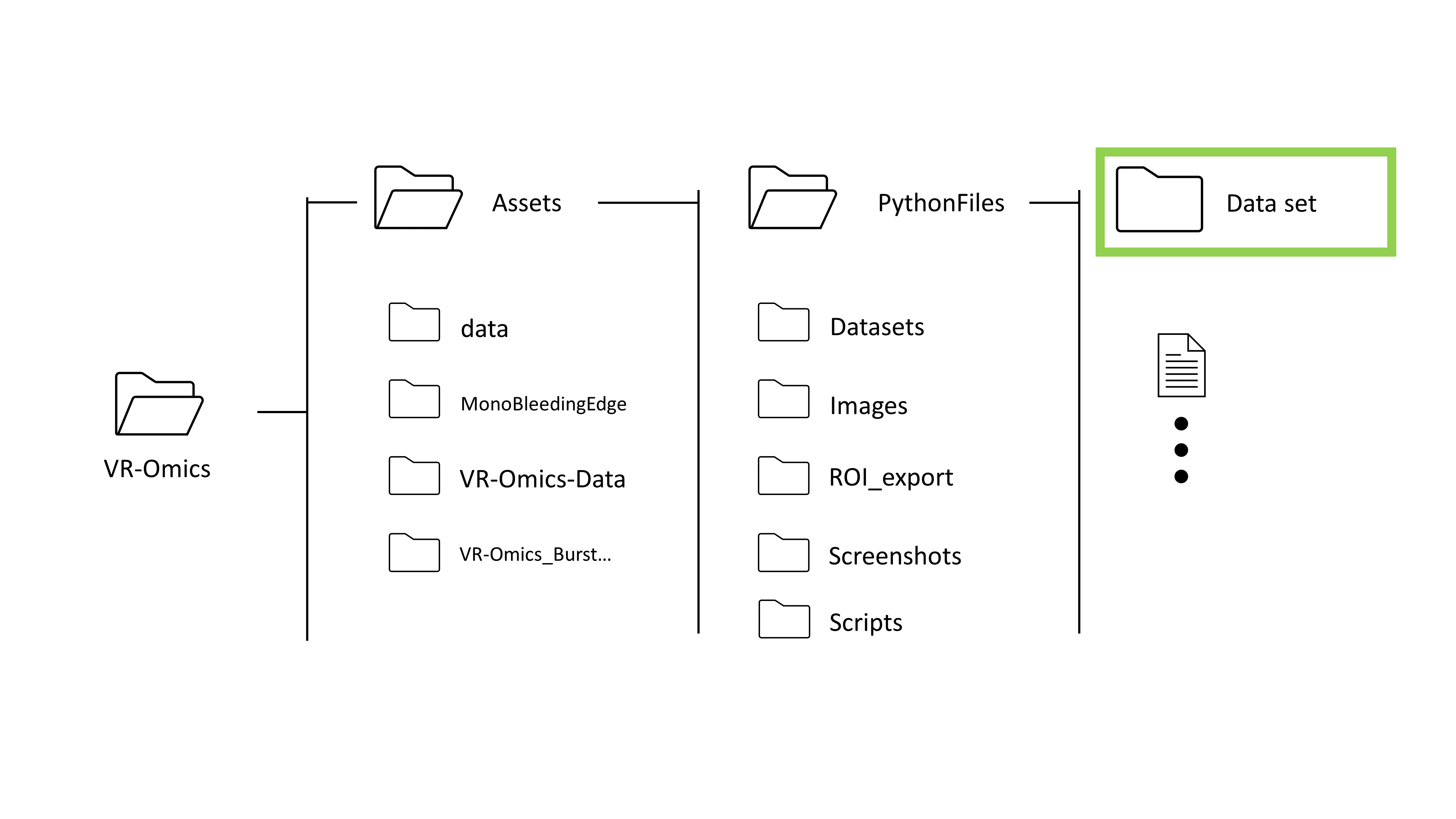
|
Load data from local machine
Loading a single Visium slide
| 1 | Load a single Visium dataset |
|

|
|---|---|---|---|
| 2 | Select sample folder |
|
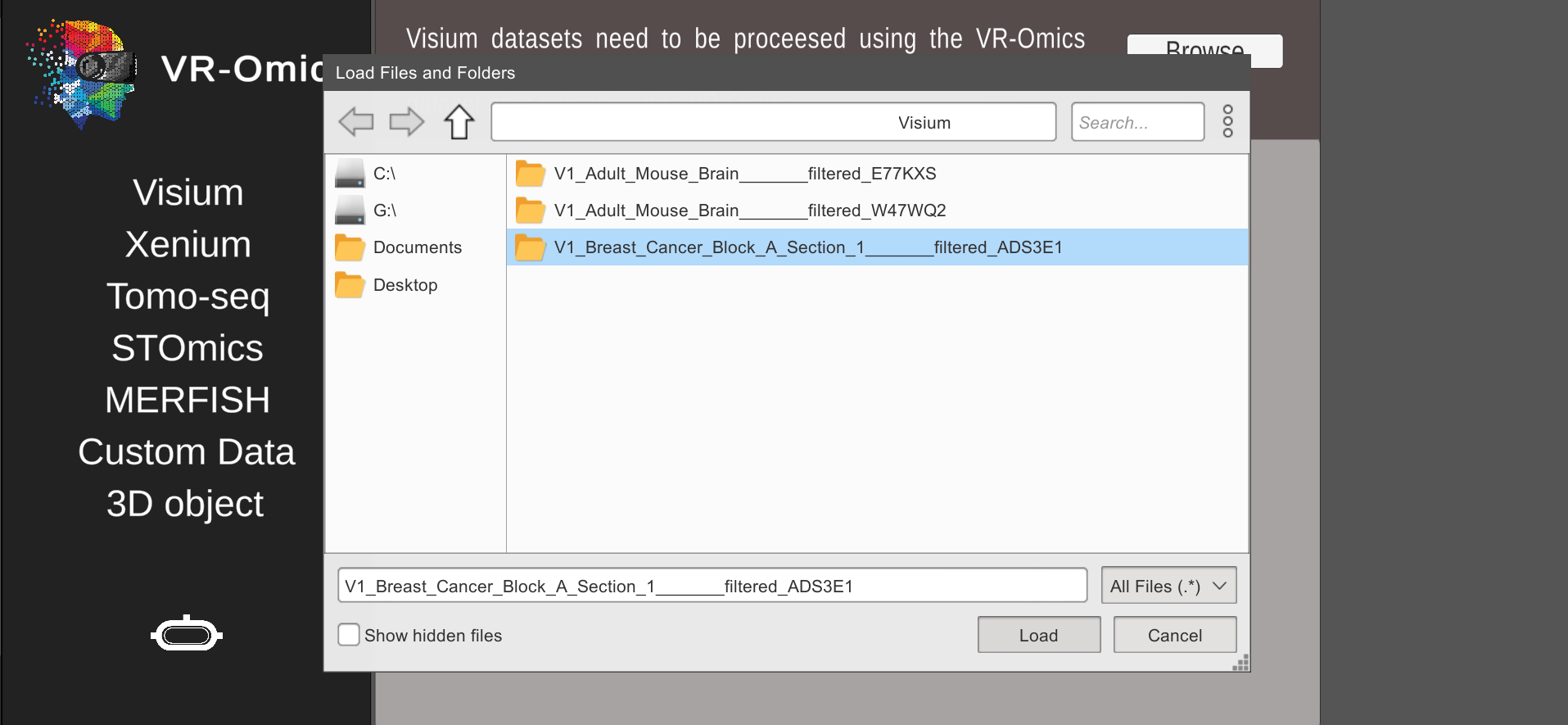

|
| 3 | Confirm selection |
|
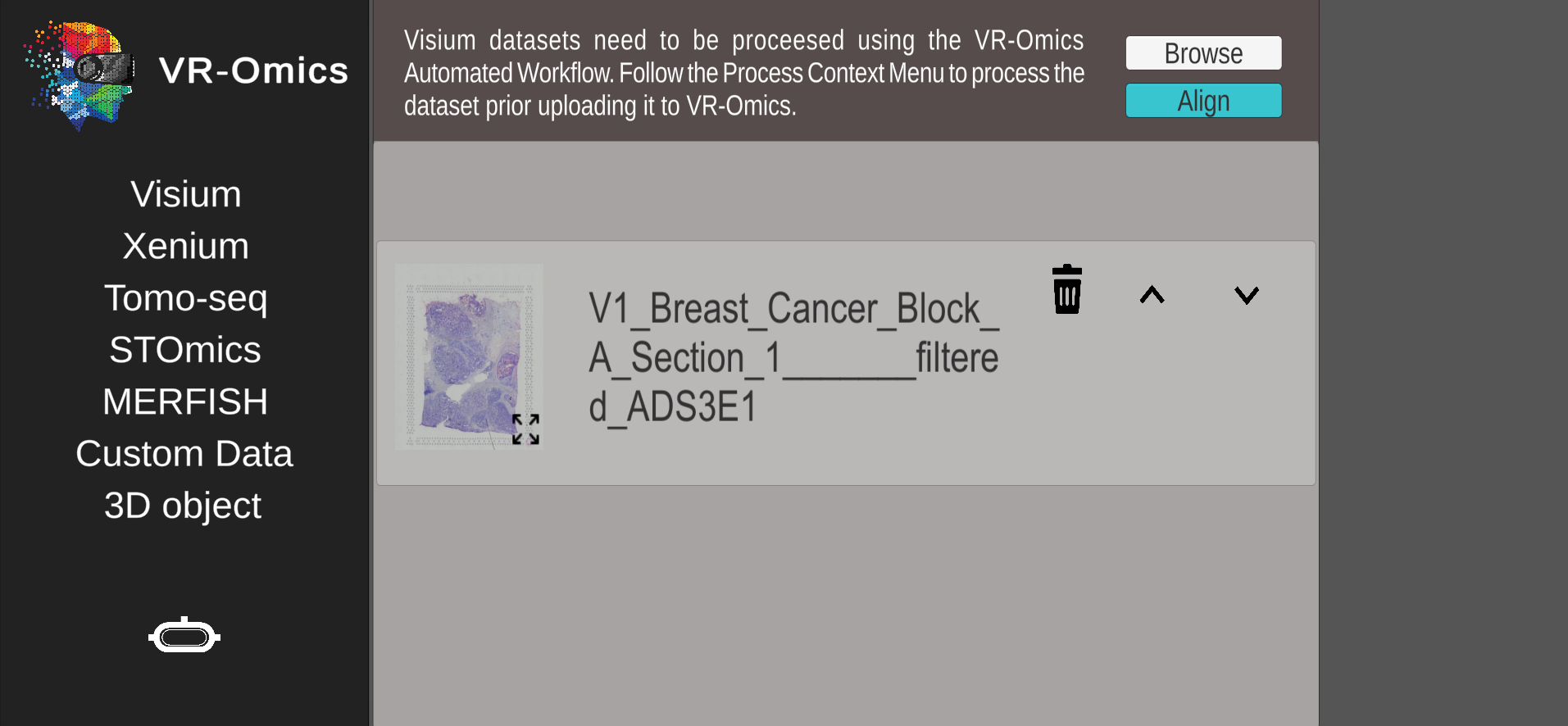
If the tissue image wasn't found a default VR-Omics placeholder will be shown. To fix this, please ensure that the respective file within the sample folder is named The dataset can be visualised and explored without using the tissue image with some limited functionalities. |
| 4 | Rotate the samples in the 3D mapping tool |
|
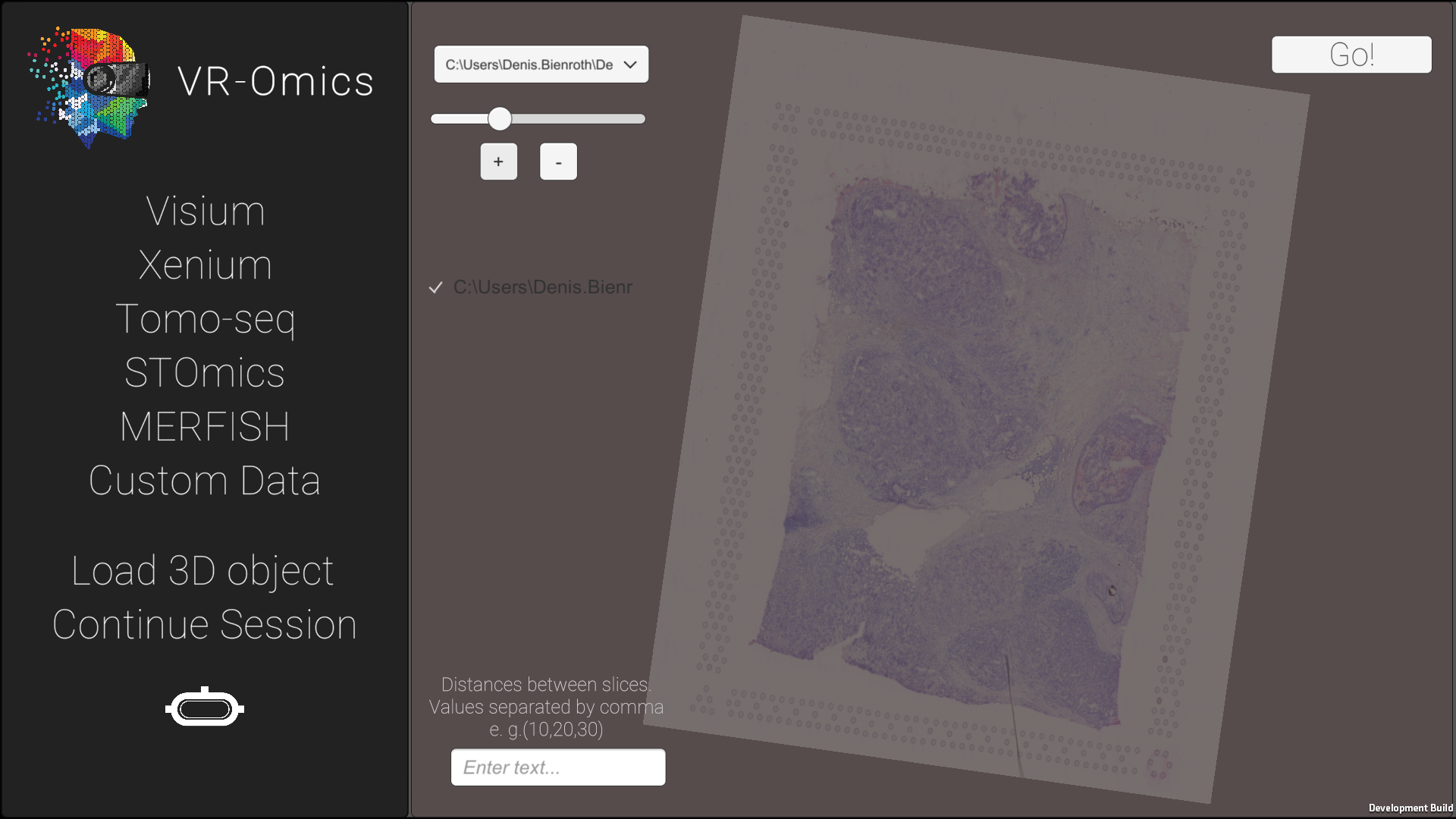
|
Loading multiple Visium slides in 3D
| 1 | Load multiple Visium datasets in 3D |
|

|
|---|---|---|---|
| 2 | Select sample folders |
|
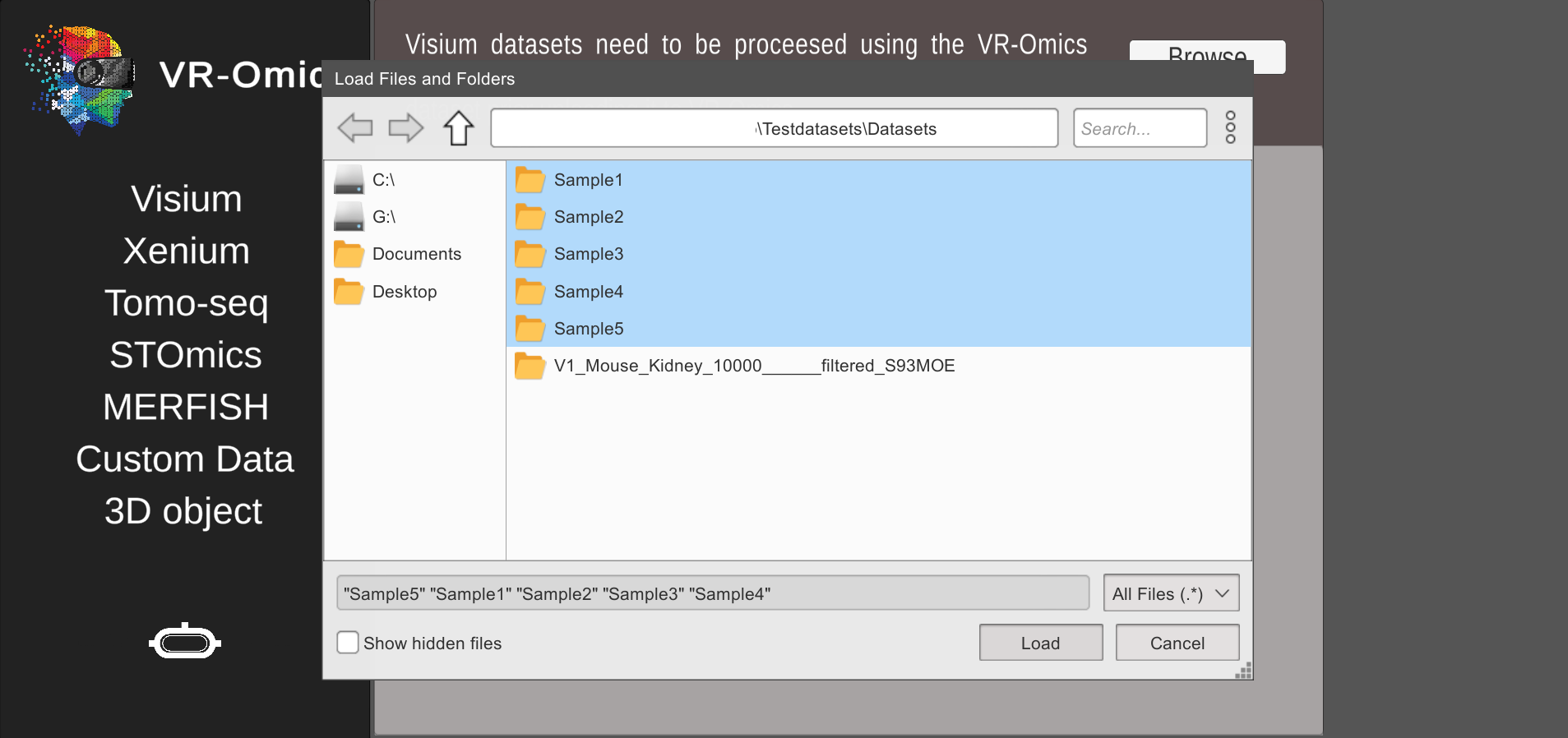

../VR-Omics/Assets/PythonFiles/sample/ |
| 3 | Confirm selection |
|
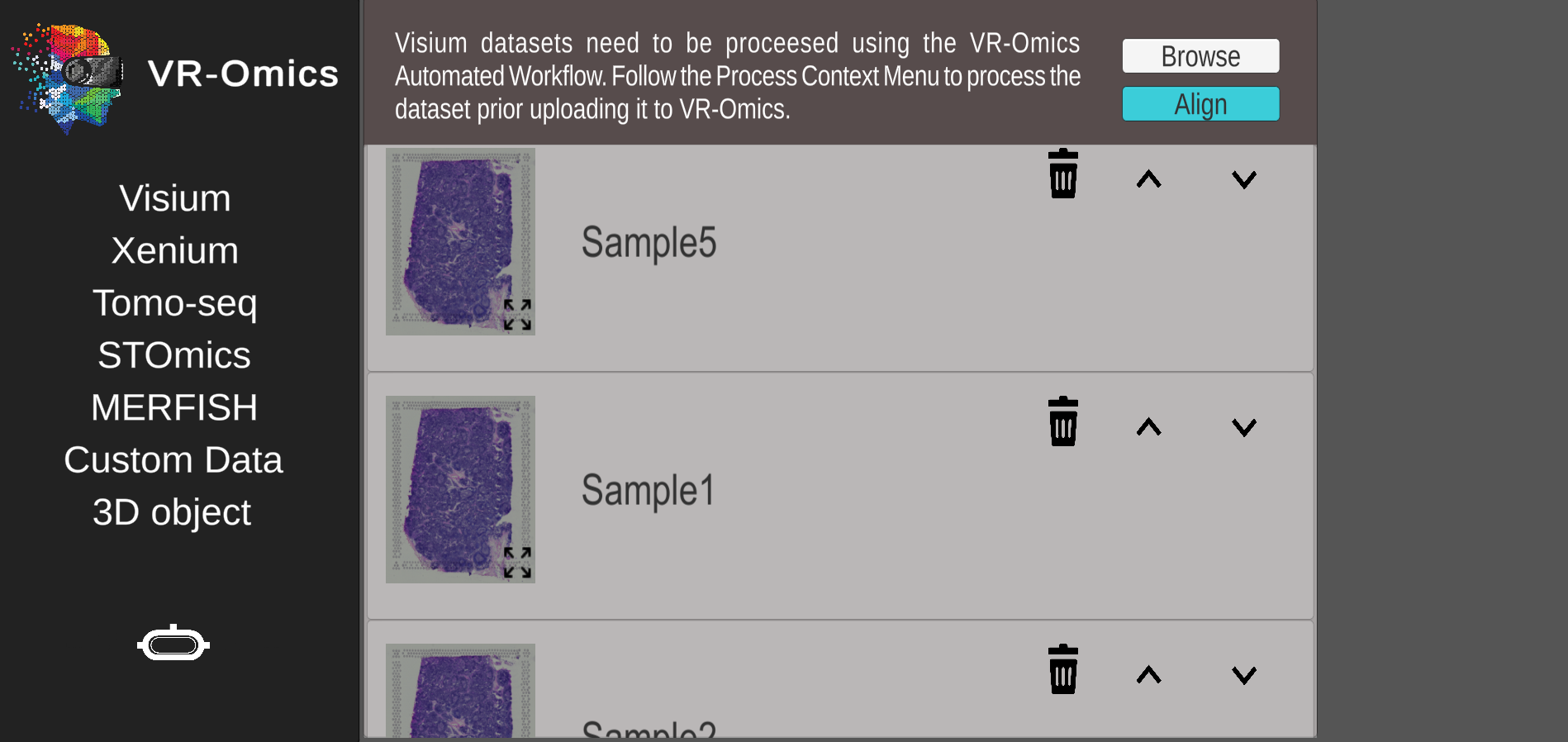
If the tissue image wasn't found a default VR-Omics placeholder will appear. To fix this, please ensure that the respective file is named |
| 4 | Rotate the samples in the 3D mapping tool |
|
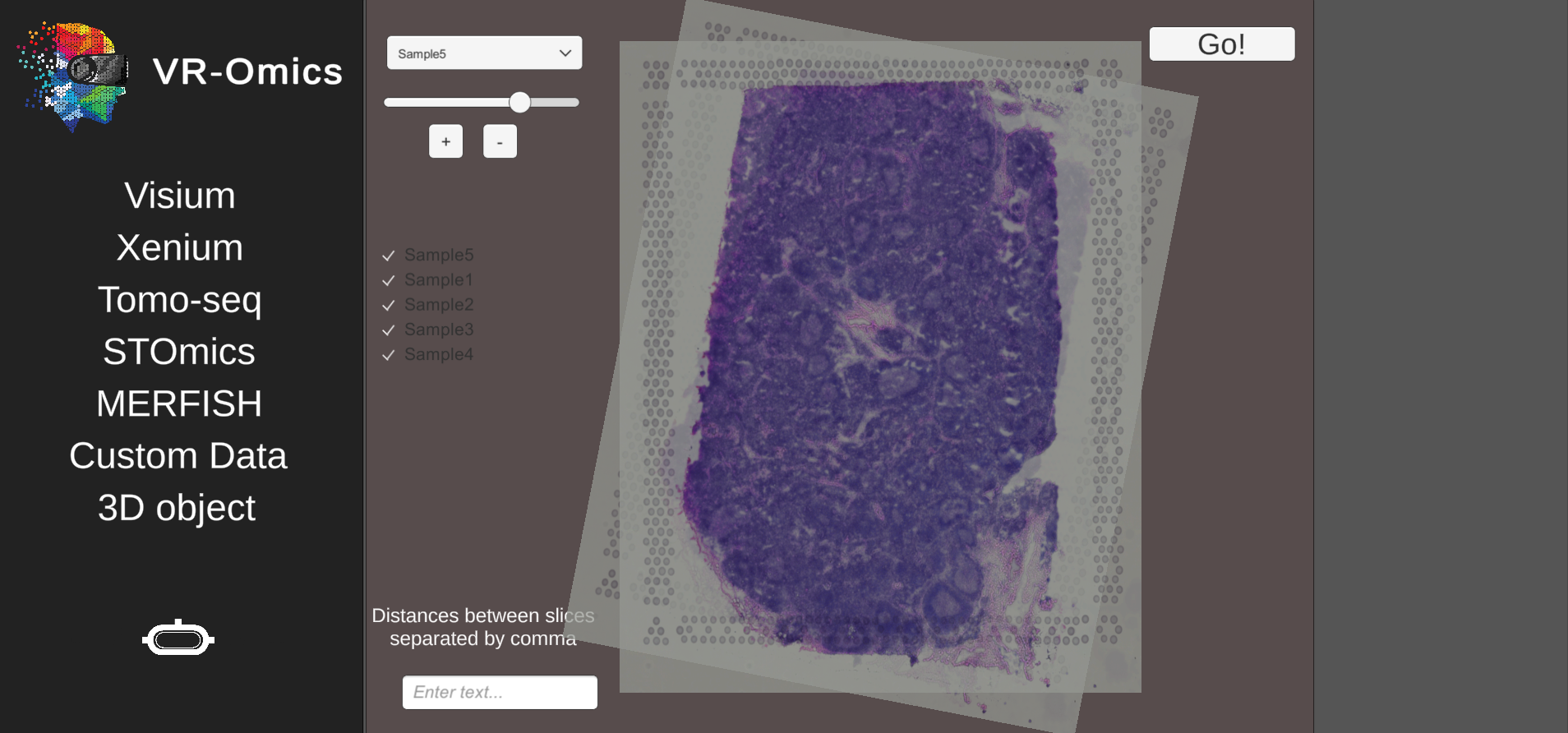
|
Process
| 1 |
Process Visium dataset from local machine Combinable with: None |
|

|
|---|---|---|---|
| 2 |
Process Visium dataset from local machine Combinable with: None |
|
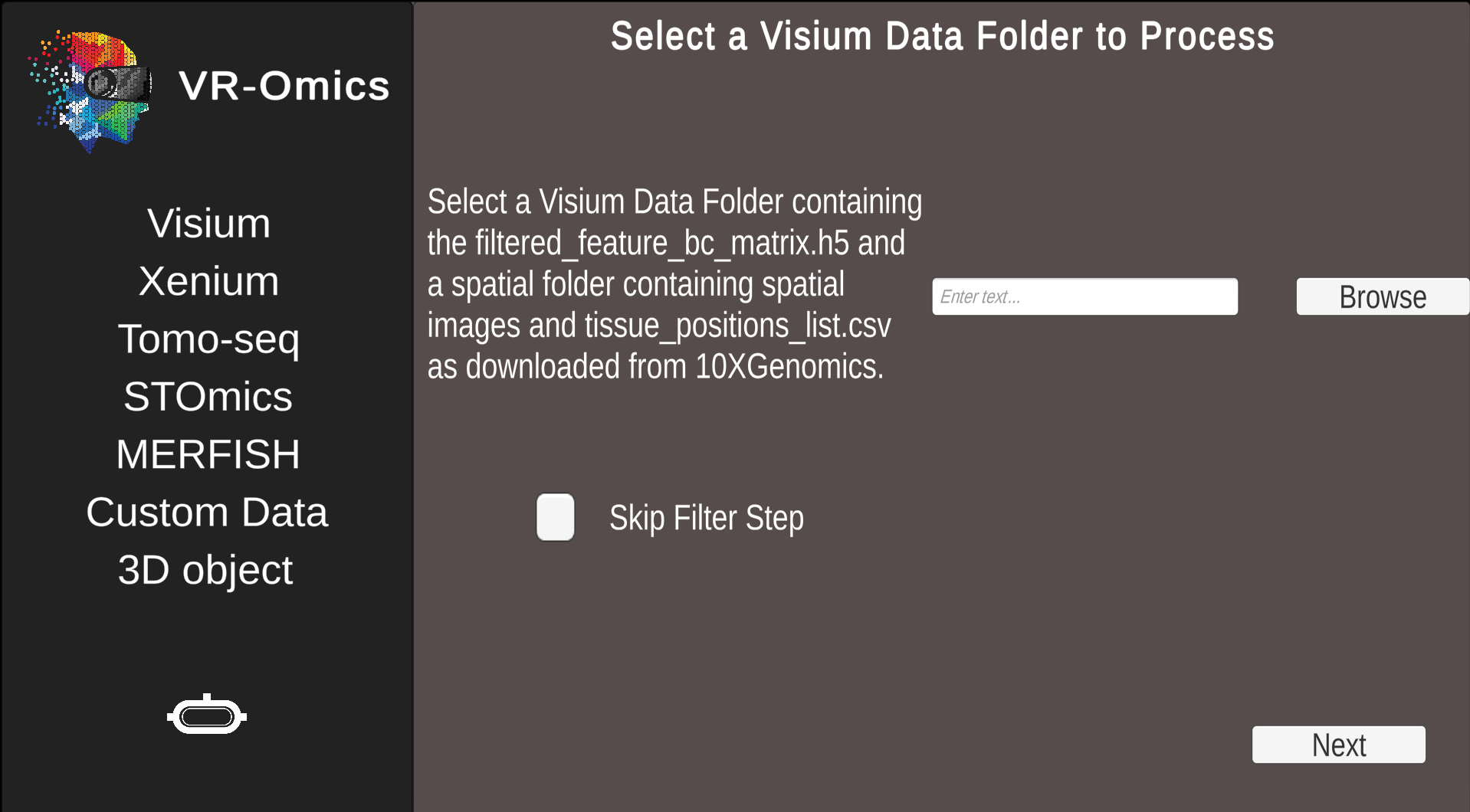
filtered_feature_bc_matrix.h5 |
| 3 |
Process and Filter Visium dataset from local machine Combinable with: Spatially Variable Genes |
|


To modify the p-value, go The samples processed using the VR-Omics workflow are stored in the VR-Omics application folder on your local machine. 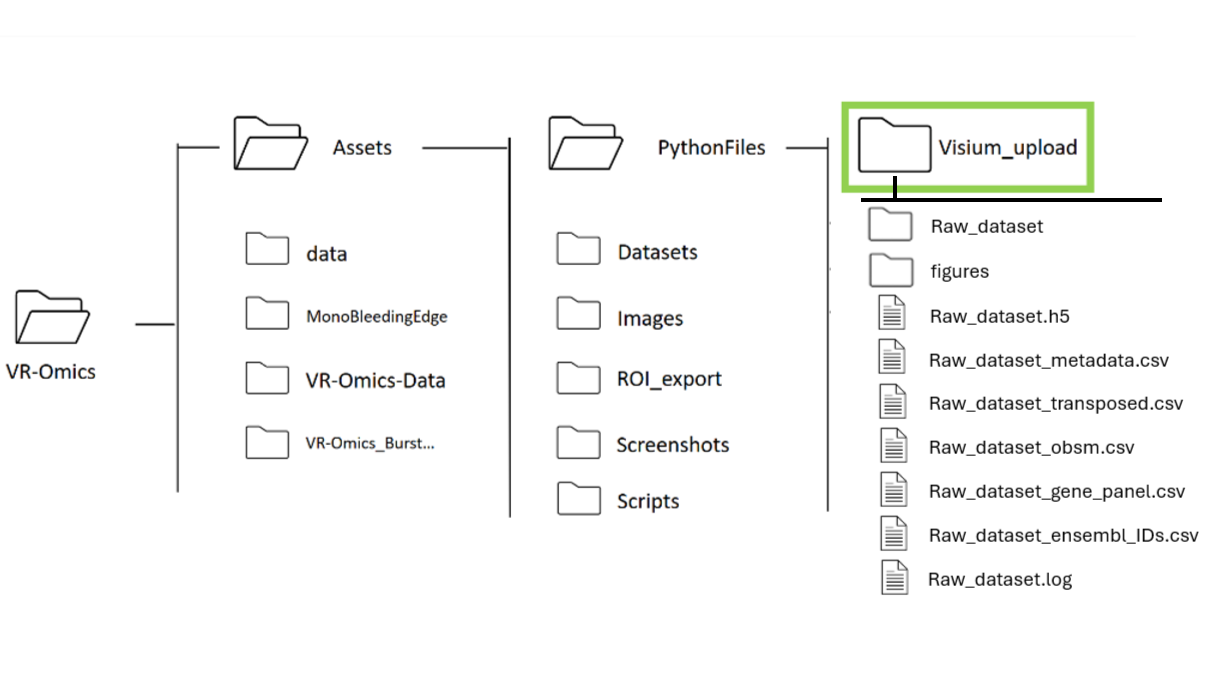
After clicking Process, the Python executable will be started. Don't close the pop up windows and wait until the pipeline has finished. This might take a couple of minutes. |
Concat
| 1 | Navigate to the Visium concat menu |
|

|
|---|---|---|---|
| 2 | Select datasets to be aligned |
|

|
3 | Check the selected samples |
|
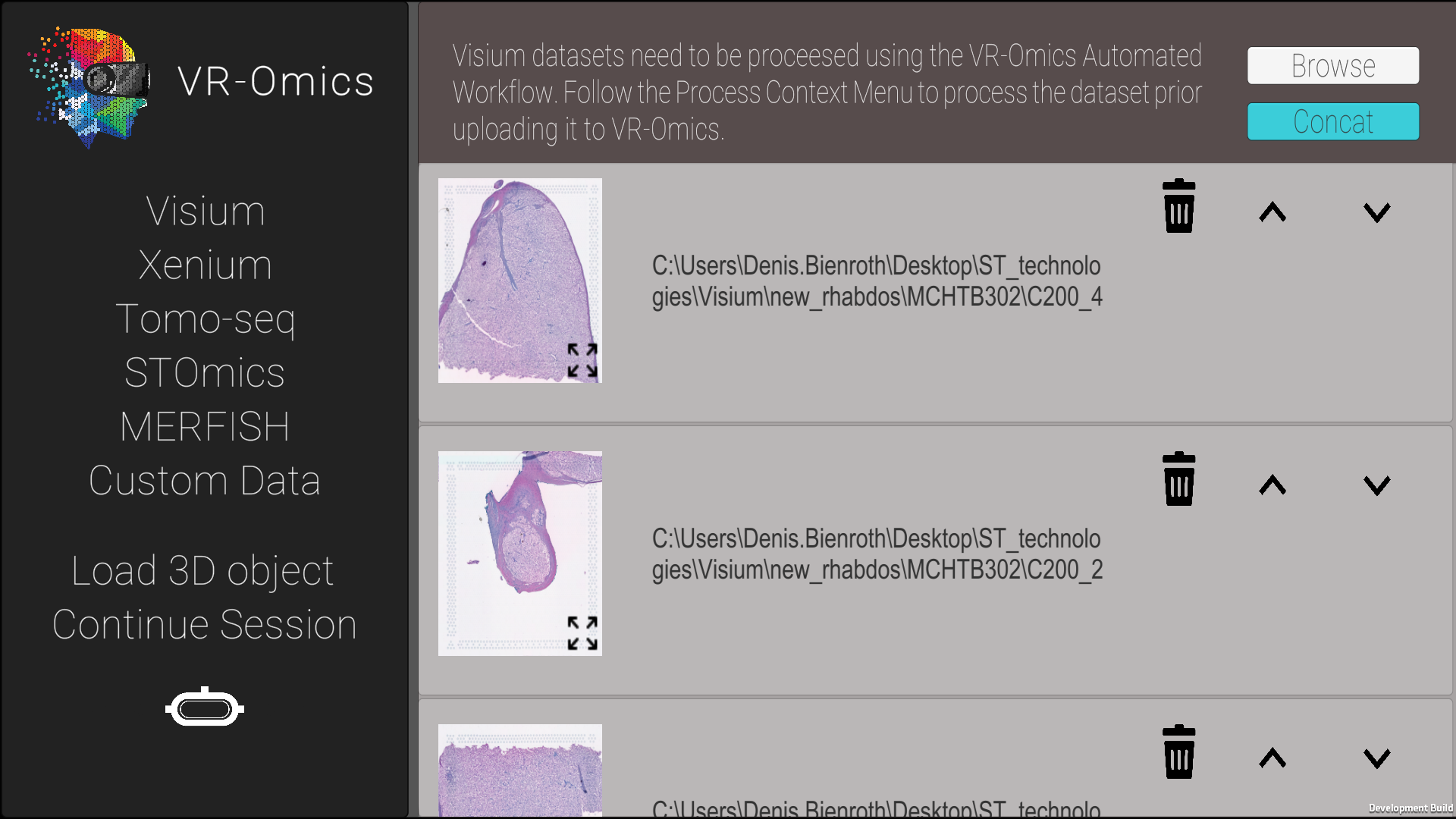
|
| 4 | Arrange the slides |
|
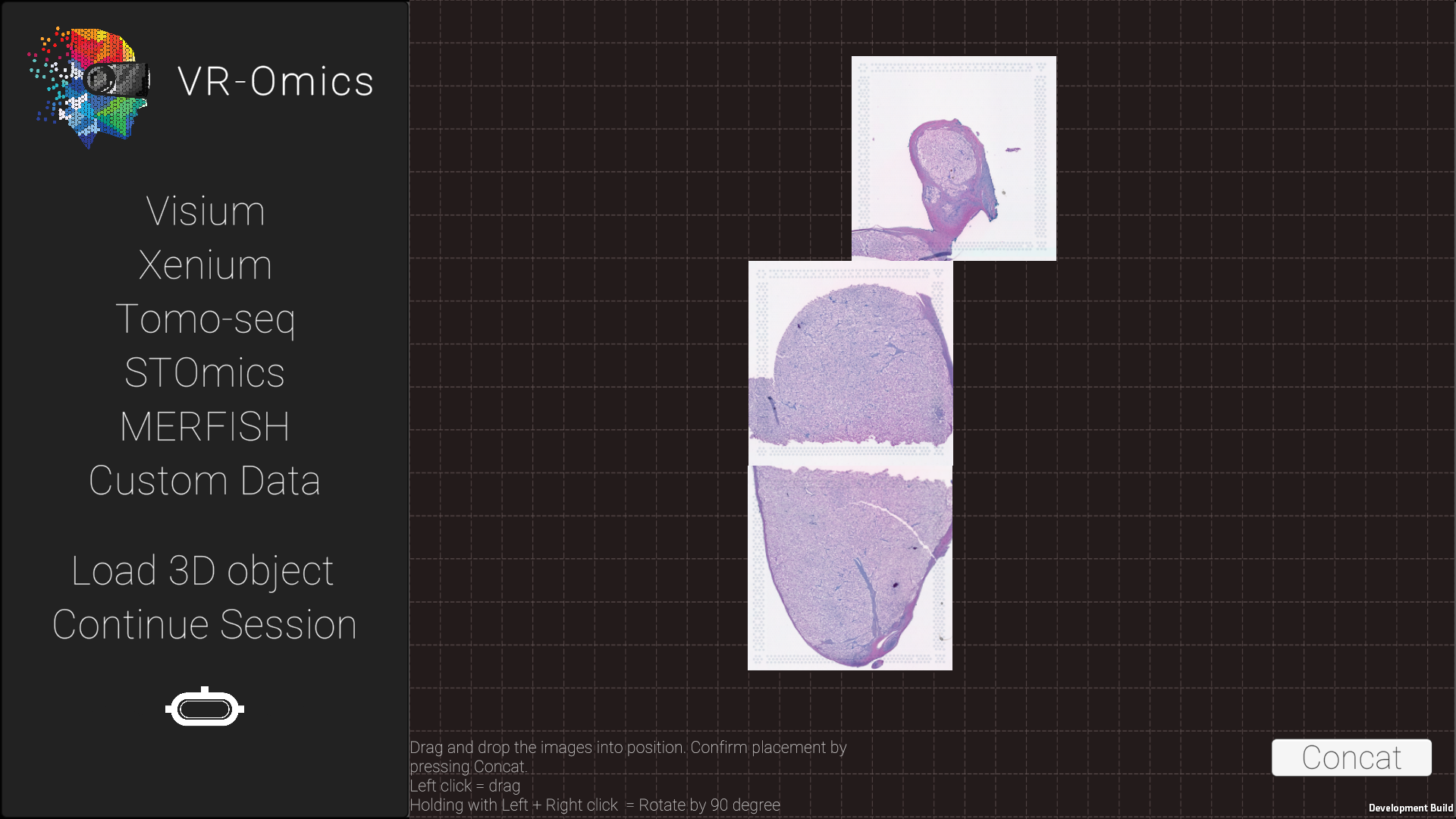
|
| 5 | Processing the joint dataset |
|
The output will be saved in the VR-Omics folder on your local machine. 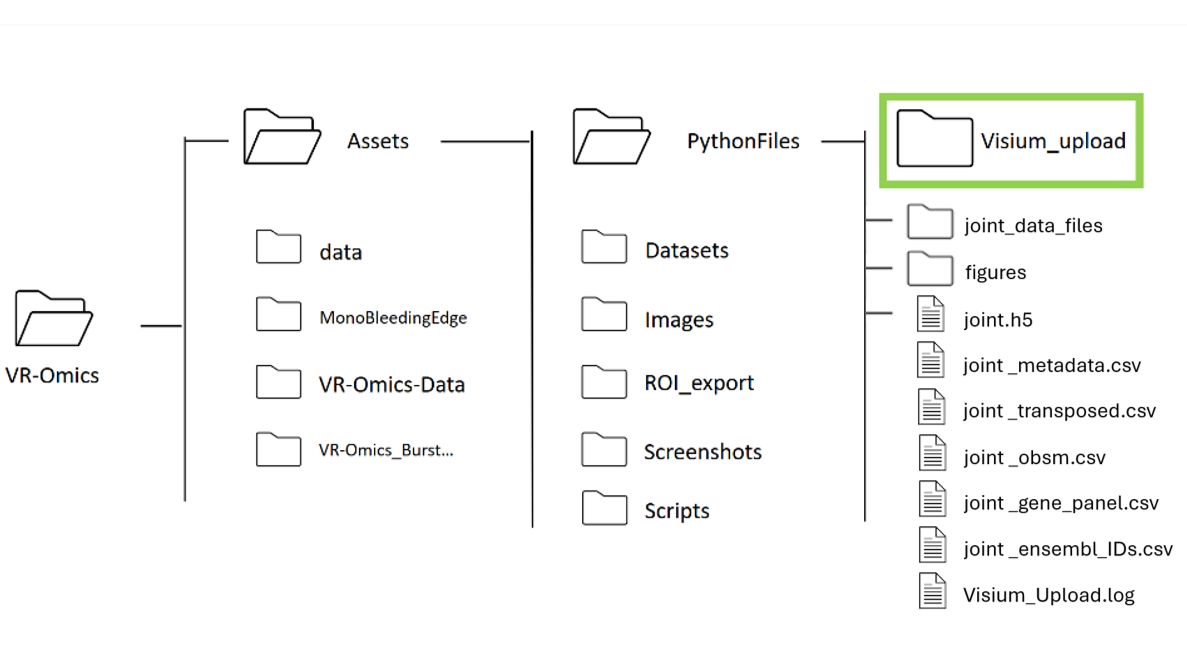
|
Demo
| 1 | Explore the Demo dataset |
|

|
|---|---|---|---|
| 2 | Demo in Visualiser |
This will automatically load the demo dataset in the VR-Omics Visualiser ready to be explored. |
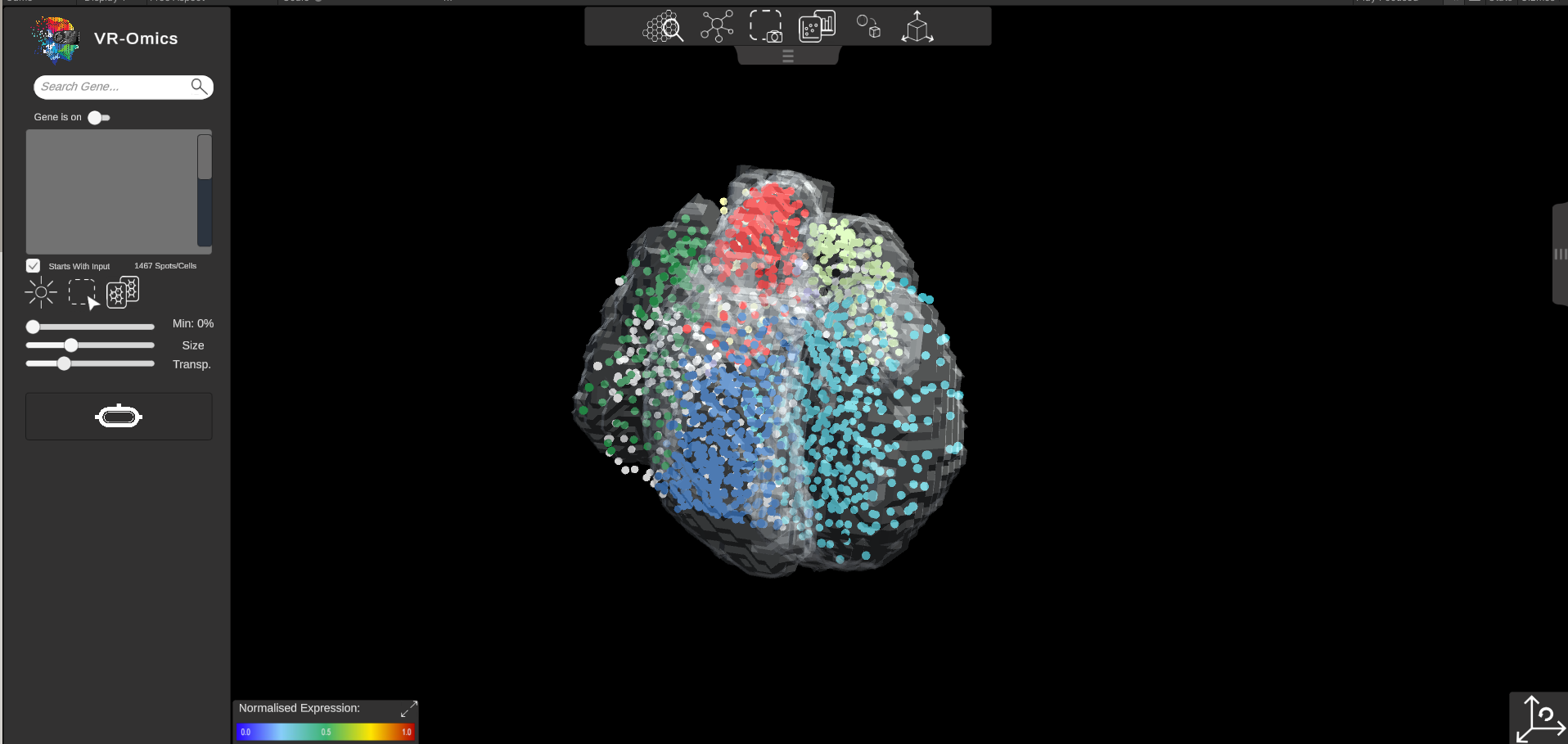
|
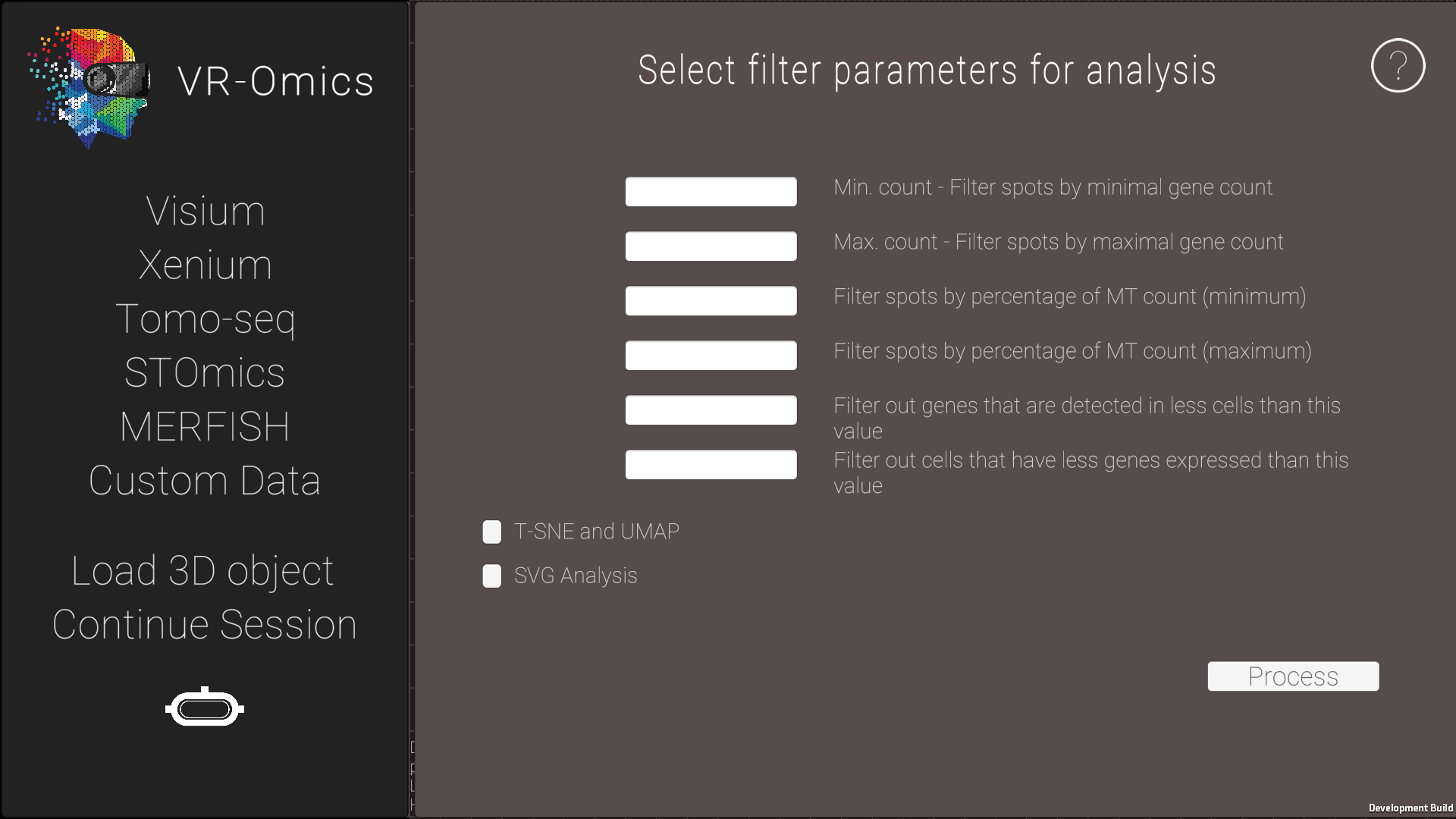
Filter parameters
|
Min.count - Filter spots by gene count |
Min.count is a filter used to exclude spots with gene expression lower than a specified value from further analysis. It is used to ensure more accurate and robust results in gene expression experiments. |
|
Max.count - Filter spots by gene count |
Max.count is a filter used to exclude spots with gene expression Lower than a specified value from further analysis. It is used to ensure more accurate and robust results in gene expression experiments. |
| Filter spots by percentage of MT count (minimum) | MT.count (minimum) is the filter parameter used to exclude spots that have a lower Mitochondrial count than the specified value. |
| Filter spots by percentage of MT count (maximum) | MT.count (maximum) is the filter parameter used to exclude spots that have a Lower Mitochondrial count than the specified value. A Low mitochondria count can be a sign of cell death or cell damage or stress. |
| Filter out genes that are detected in less spots than this value | This minium threshold filter parameter can be used to exclude all genes from the dataset that are detected in less spots than the value provided. Keep in mind the total number of genes of the dataset. VR-Omics will skip this step if this value was selected to Low so that no genes remain or if the value exceeds the total genes number of the dataset. |
| Filter out spots that have less genes expressed than this value. | This minium threshold filter parameter can be used to exclude all spots from the dataset that have less genes expressed than this value. Please keep the maximum number of spots in the dataset in mind. VR-Omics will skip this step if the value was selected so Low, that no spots would remain after analysis. |
Further information can be found in the Scanpy spatial analysis documentation.
Required Folder Format for samples not processed with VR-Omics
For samples not downloaded or processed using the VR-Omics Automated Workflow, ensure this folder structure. Further descriptions of the files can be found below. Alternatively, they can be uploaded using the custom data option with limited features.
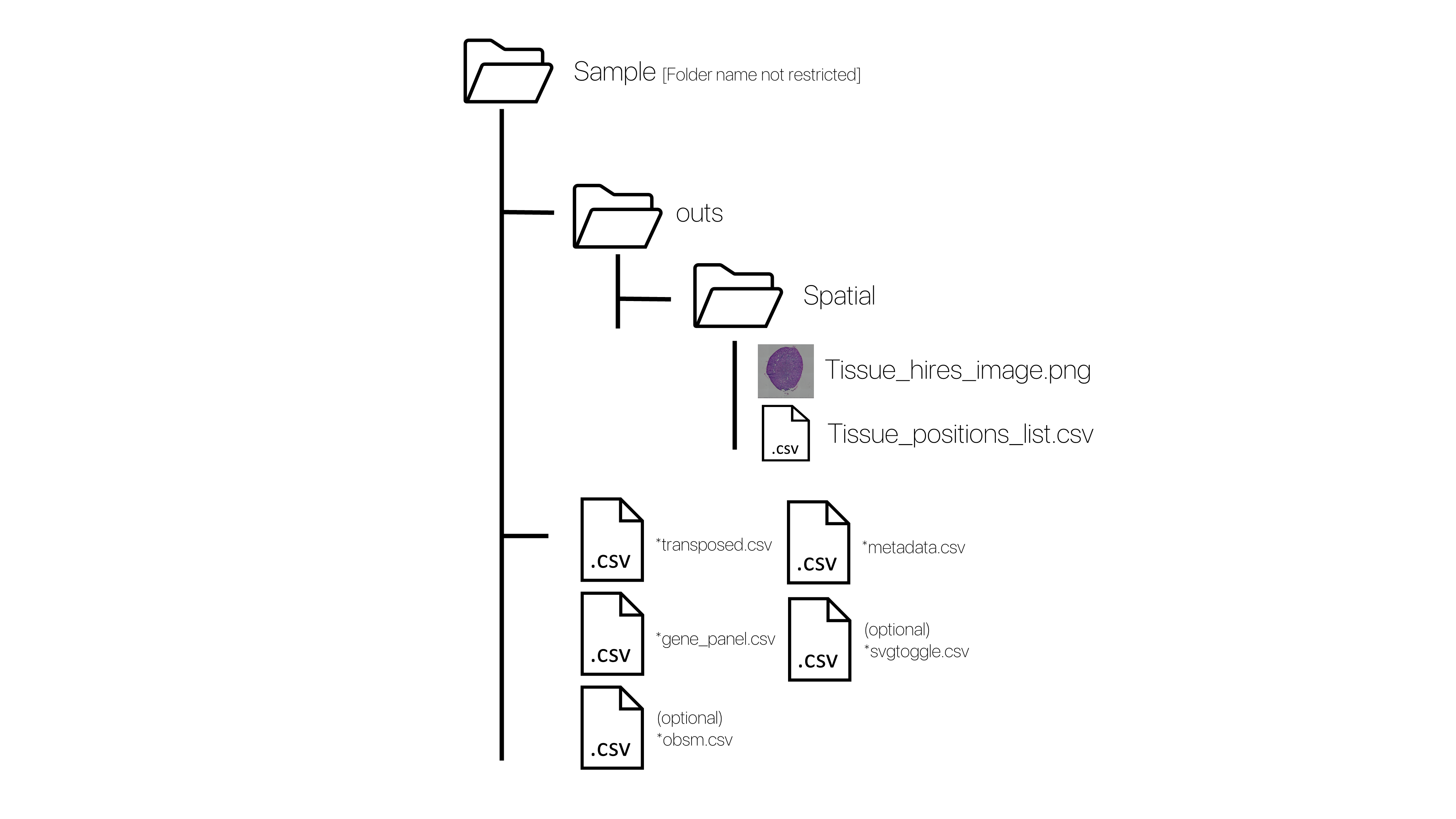
| Required file name ending | Full name (VR-Omics) | Description | Required |
|---|---|---|---|
tissue_hires_image.png
|
tissue_hires_image.png
|
The high-resolution tissue image (2000 x 2000 pixels) as provided by 10x genomics. | false but impacts features |
Tissue_positions_list.csv
|
Tissue_positions_list.csv
|
CSV file as provided by 10x genomics. File does not contain headers. 1. Spot IDs; 2. spot on tissue; 3. + 4. row and column of spot; 5. + 6. X and Y coordinates of spot; | true |
transposed.csv
|
outs_transposed.csv
|
CSV file containing gene counts per spot. Header contains spot IDs. First column contains gene names. Each row contains gene count information per spots (columns). | true |
metadata.csv
|
outs_metadata.csv
|
Header: in_tissue, array_row, array_col, n_genes_by_counts, log1p_n_genes_by_counts, total_counts, log1p_total_counts, pct_counts_in_top_50_genes, pct_counts_in_top_100_genes, pct_counts_in_top_200_genes, pct_counts_in_top_500_genes, total_counts_mt, log1p_total_counts_mt, pct_counts_mt, n_counts, n_genes, clusters
|
true |
gene_panel_IDs.csv
|
outs_gene_panel_IDs.csv
|
CSV file with one column header name gene listing all gene names in the dataset.
|
true |
svgtoggle.csv
|
outs_svgtoggle.csv
|
g, FSV, M, g, l, max_delta, max_ll, max_mu_hat, max_s2_t_hat, model, n, s2_FSV, s2_logdelta, time, BIC, max_ll_null, LLR, pval, qval
|
false |
obsm.csv
|
outs_obsm.csv
|
1. column spot IDS. 2. + 3. Spot coordinates in pixels. Followed by a variable number of columns for X_PCA values, UMAP X and Y coordinates, and TSNE X and Y coordinates. | false |
Xenium
Process
| 1 | Navigate to the Xenium process menu |
|

|
|---|---|---|---|
| 2 | Select the Xenium files |
|
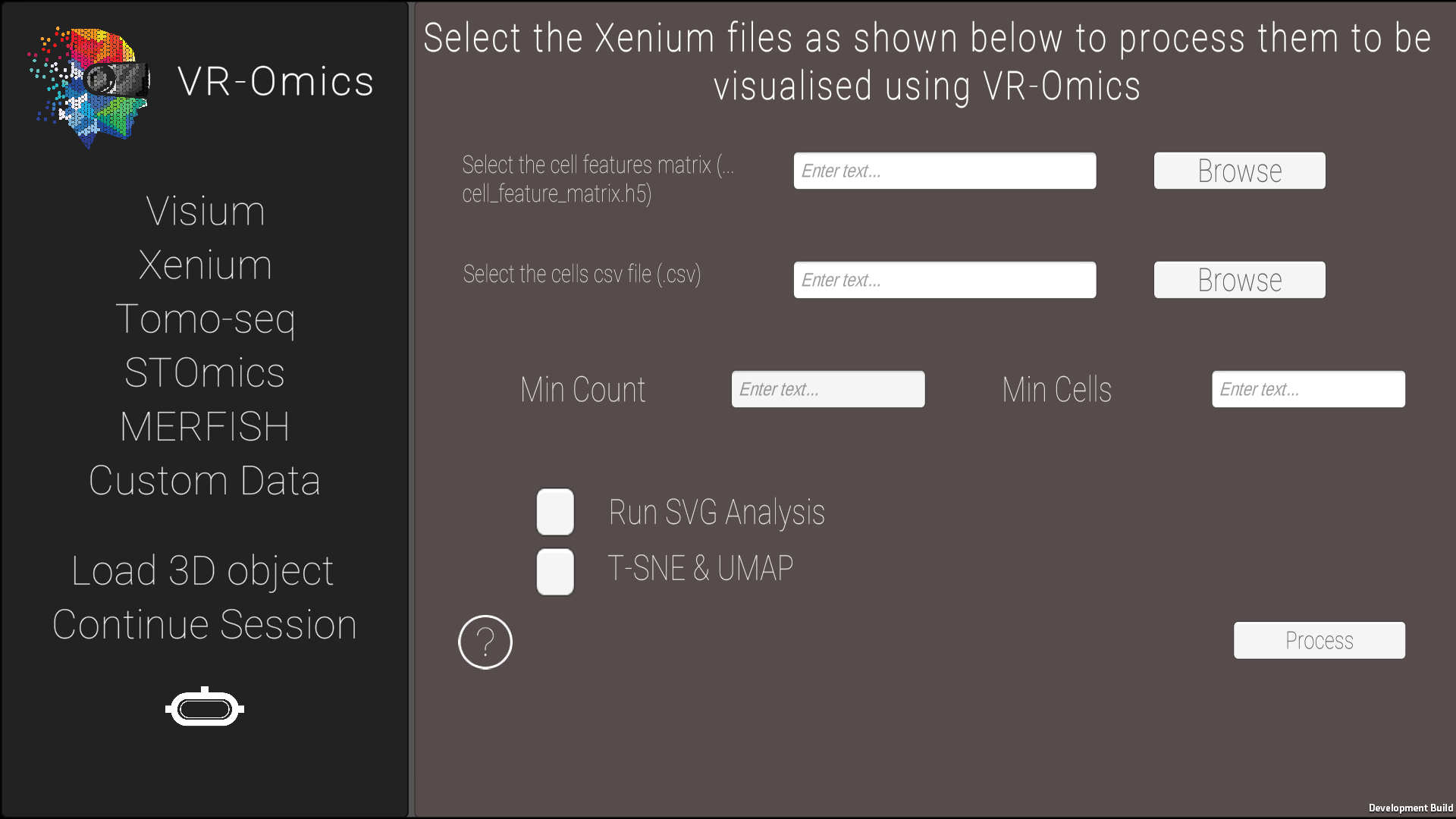
Selecting SVG Analysis and/or T-SNE/UMAP will increase processing time of the sample. 
p-value threshold for SVG cutoff can be customized. To modify the p-value, go The processed data will be saved in the VR-Omics data folder. To Visualise the data select the complete folder as shown in the figure 
After clicking Process, the Python executable will be started. Don't close the pop up windows and wait until the pipeline has finished. This might take a couple of minutes. |
Load
| 1 | Navigate to the Xenium load menu |
|

|
|---|---|---|---|
| 2 | Select the Xenium sample folder |
|
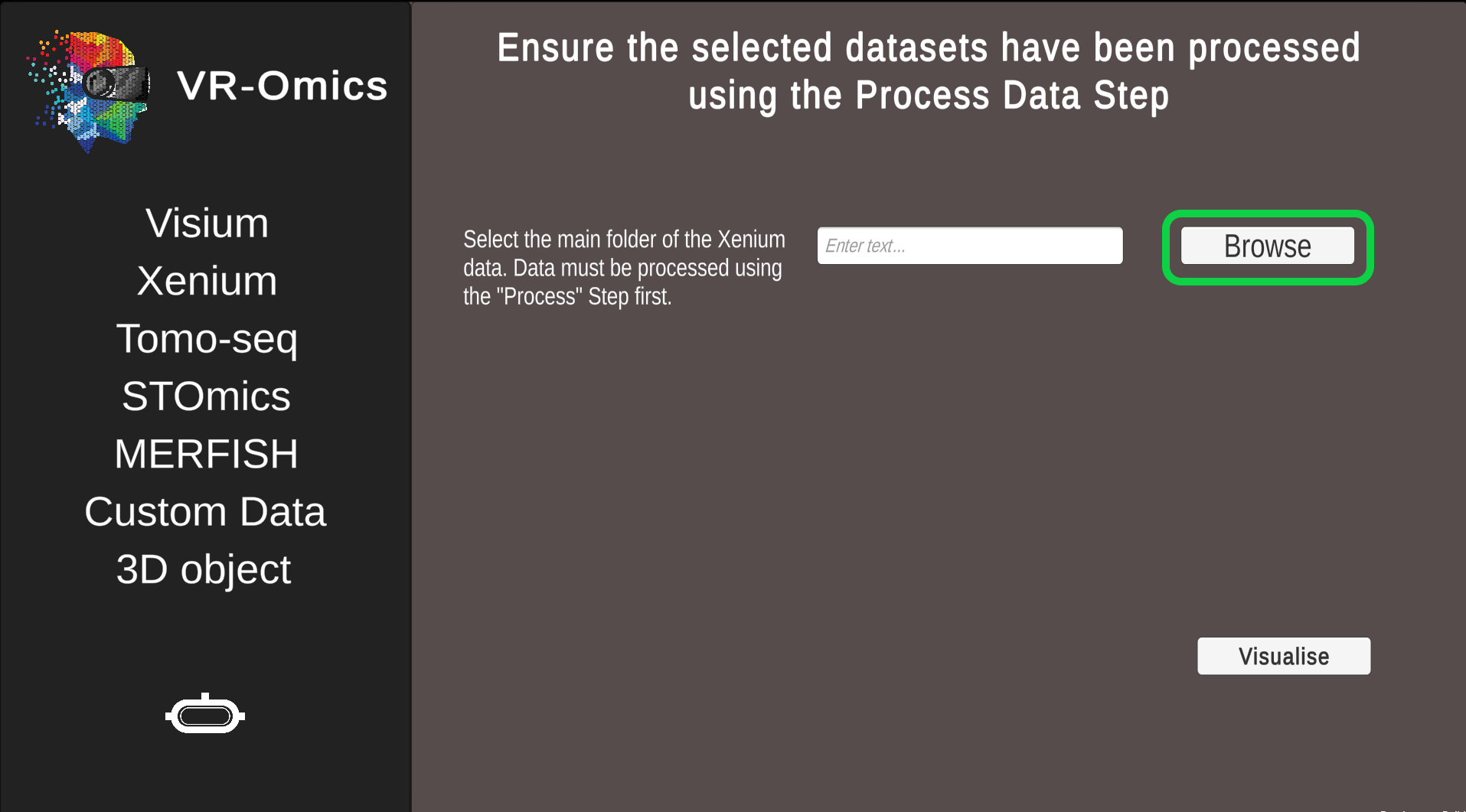
Select the Xenium folder processed with the VR-Omics AW (highlighted in green in the picture) using Browse. The data set can be found in the VR-Omics directory. Navigate from there to 
|
| 3 | Confirm selection |
Click Visualise to continue to the VR-Omics Visualiser.
|
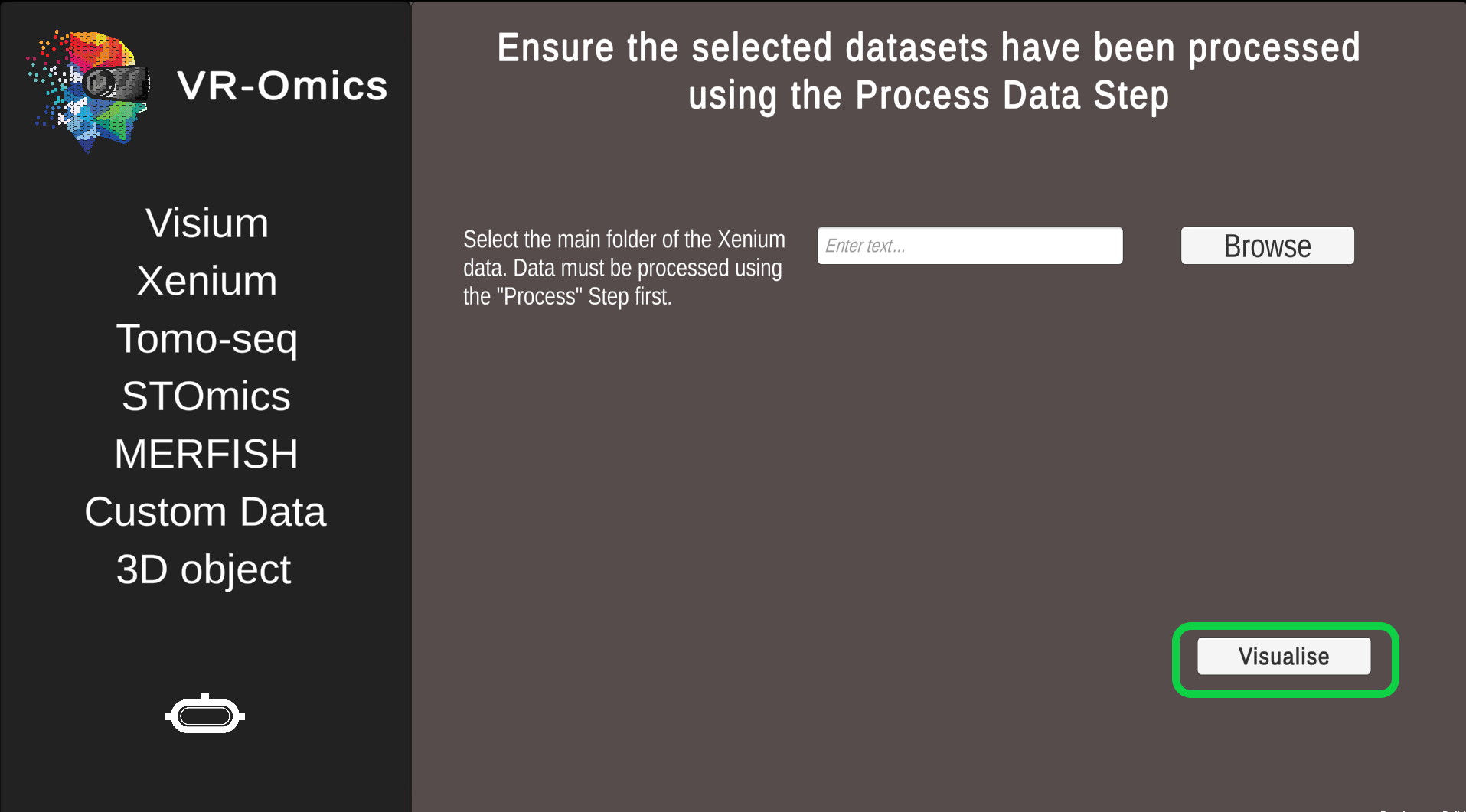
Ensure the data has been processed before visualising it using the Process function. |
Loading multiple Xenium in 3D
| 1 | Load multiple Xenium datasets in 3D |
|
|
|---|---|---|---|
| 2 | Select sample folders |
|


../VR-Omics/Assets/PythonFiles/sample/ |
| 3 | Confirm selection |
|

If the tissue image wasn't found a default VR-Omics placeholder will appear. To fix this, please ensure that the respective file is named |
| 4 | Rotate the samples in the 3D mapping tool |
|
|
| 5 | Load the 3D dataset |
|


../VR-Omics/Assets/PythonFiles/sample/ |
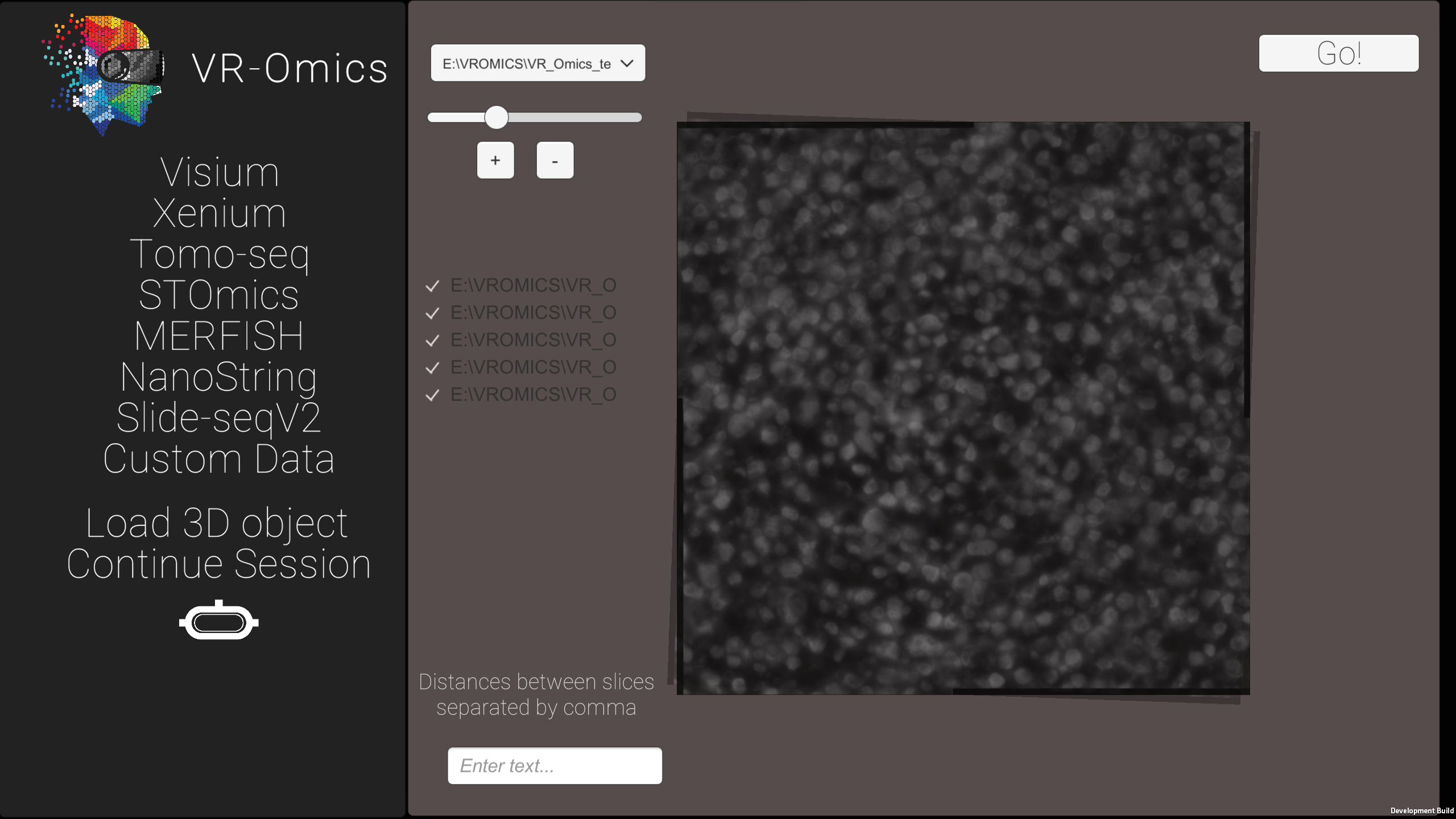
Tomo-Seq
Load
| 1 | Navigate to the Tomo-Seq load menu |
|

|
|---|---|---|---|
| 2 | Select the Tomo-Seq folder |
|
* AP = anterior to posterior, VD = ventral to dorsal, and LR = left to right The 3D reconstruction of the gene expression is explained in Asp et al. in Supplementary Data S2. |
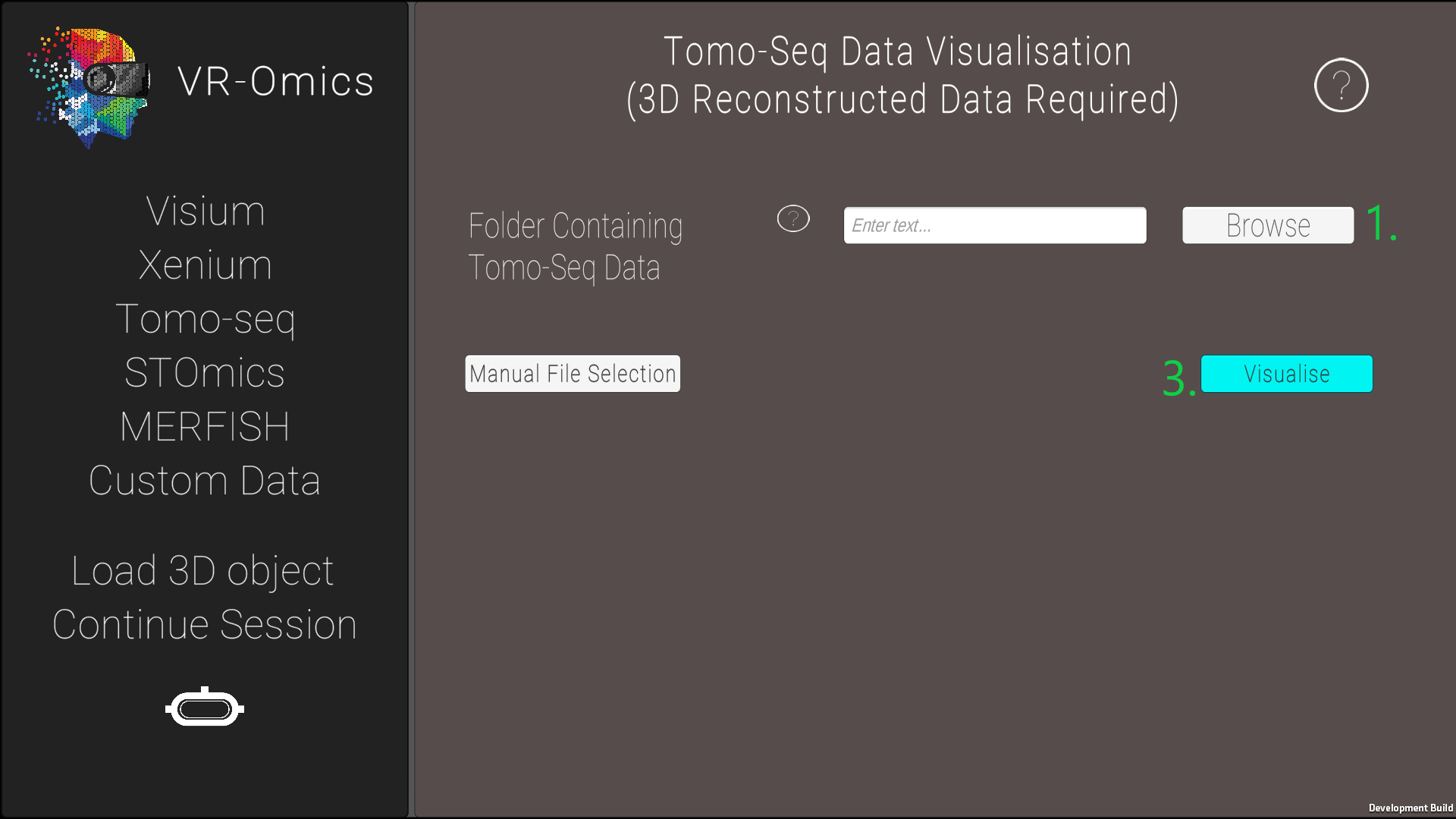
| 3 | Select the Tomo-Seq files manually |
|
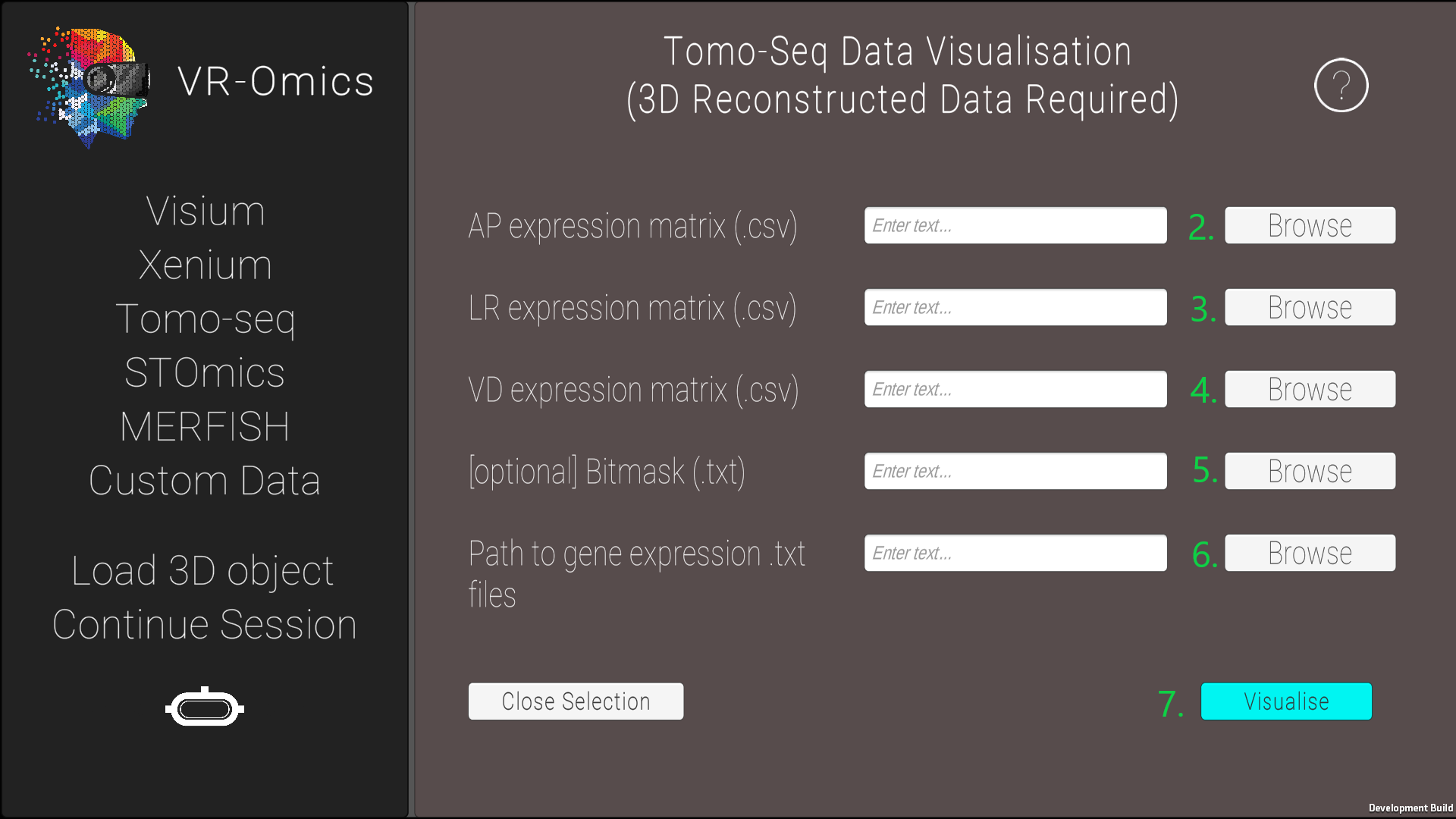
* AP = anterior to posterior, VD = ventral to dorsal, and LR = left to right The 3D reconstruction of the gene expression is explained in Asp et al. in Supplementary Data S2. |
STOmics
Process
| 1 | Navigate to the STOmics process menu |
|
|
|---|---|---|---|
| 2 | Select the STOmics file |
Select the gene expression matrix |
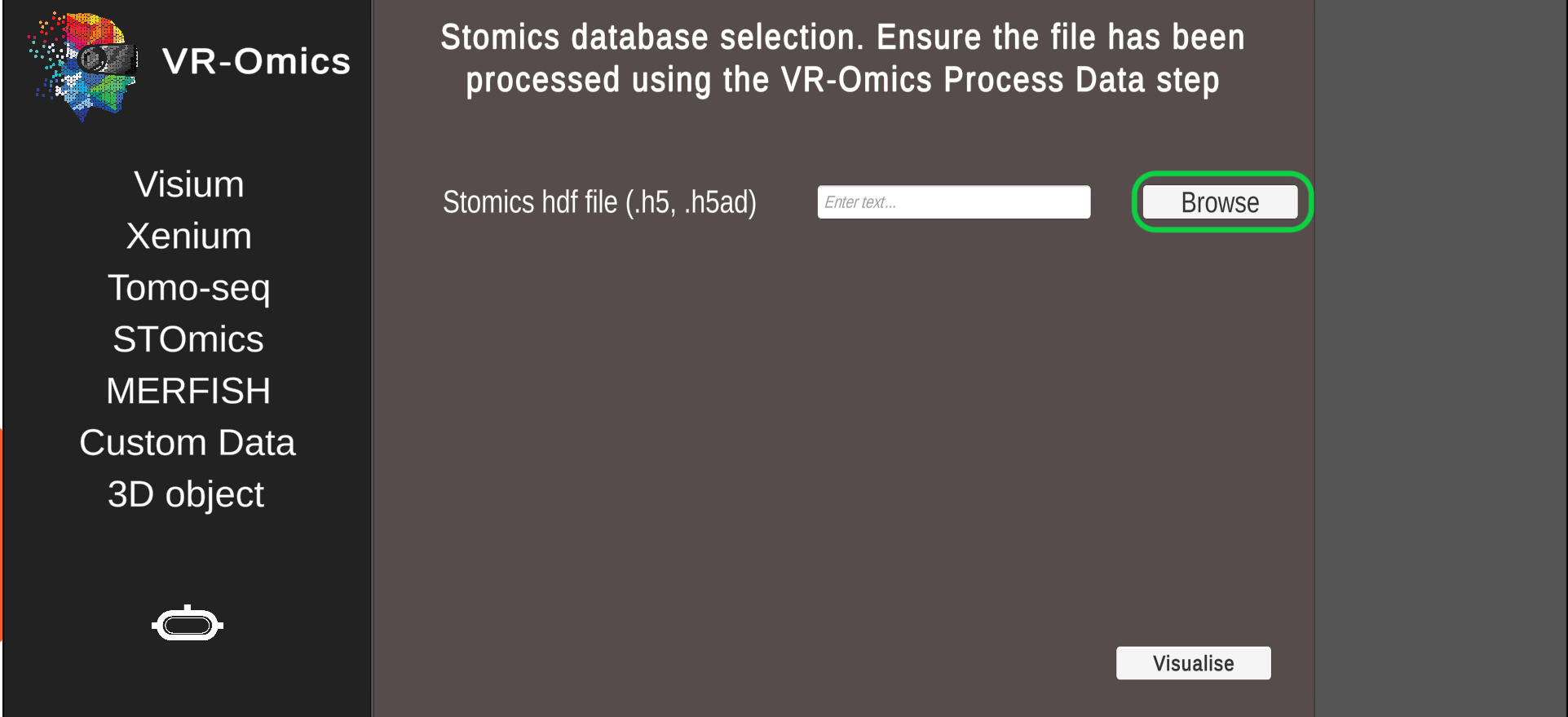
The dataset can be downloaded from STOmics Database. |
| 3 | Confirm selection |
Click Process to process the data. The processed data will be saved locally. |
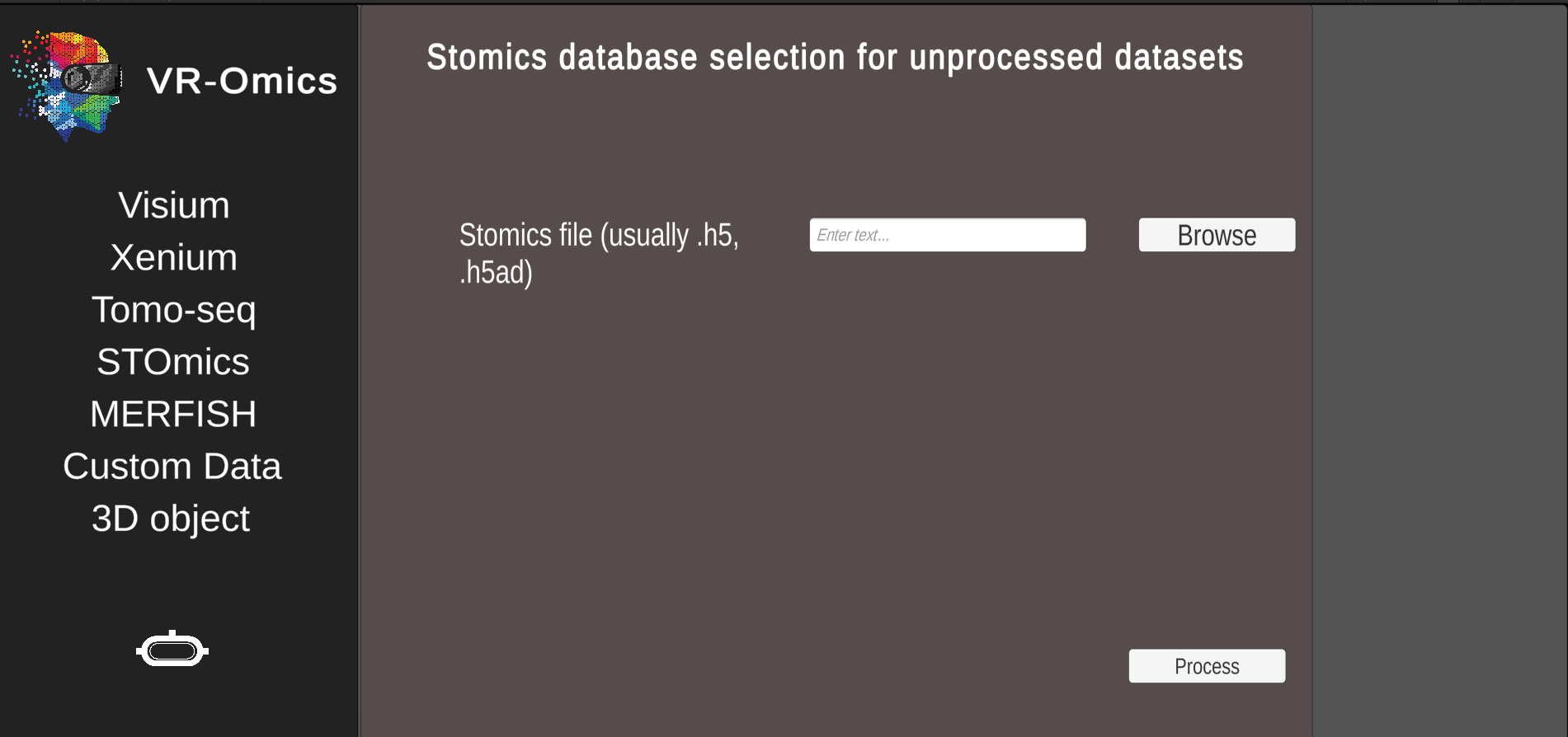
The process step can take couple of minutes depending on the size of the dataset and the computational power of the machine. Once the data was processed this step can be skipped and the data can be visualised using the STOmics load option. The processed data will be saved in the VR-Omics data folder. To Visualise the data select the complete folder as shown in the figure 
|
Load
| 1 | Navigate to the STOmics load menu |
|

|
|---|---|---|---|
| 2 | Select the STOmics files |
Select the processed gene expression STOmics |
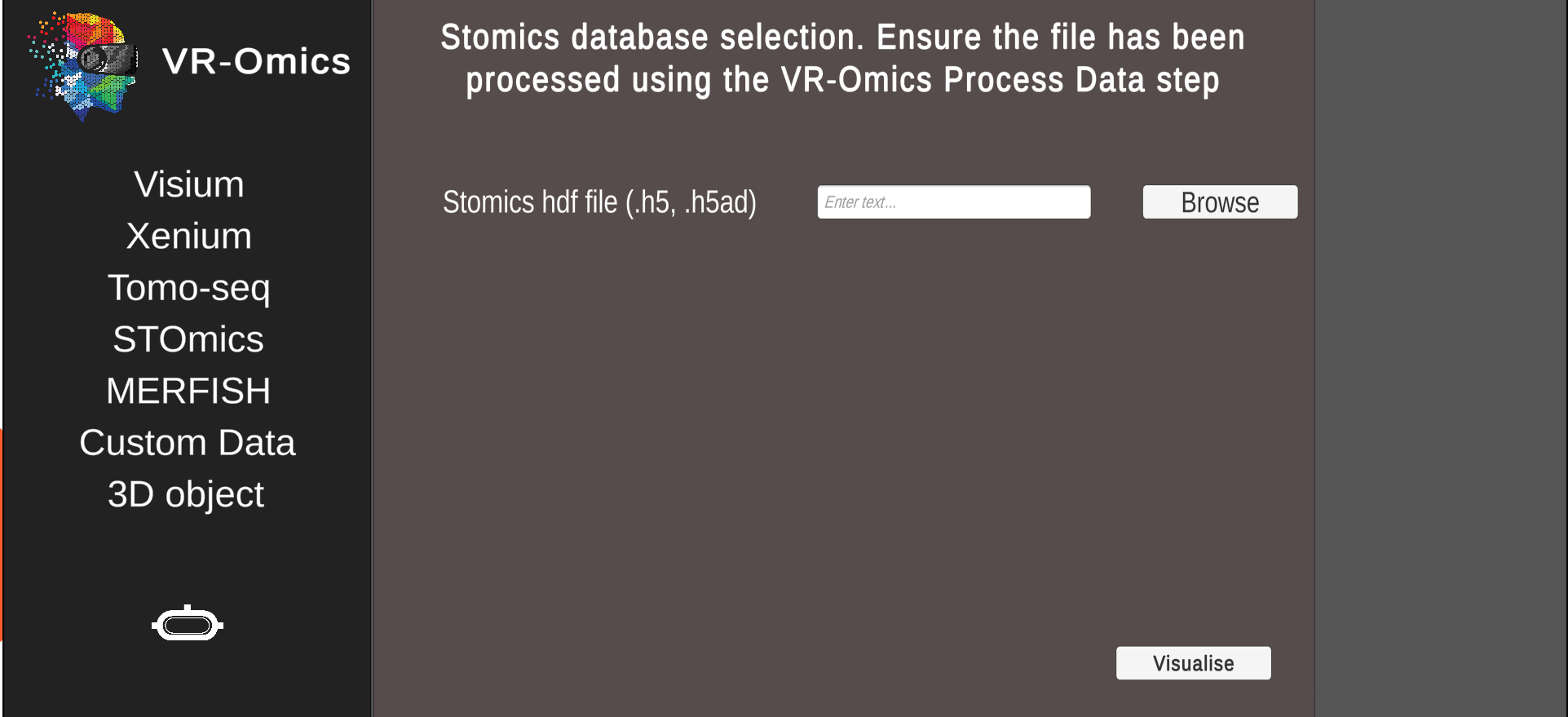
Ensure the data was processed using the Process function. The processed data can be found in the VR-Omics data folder. To Visualise the data select the complete folder as shown in the figure 
|
MERFISH
Process
| 1 | Navigate to the MERFISH process menu |
|

|
|---|---|---|---|
| 2 | Select the MERFISH files |
|
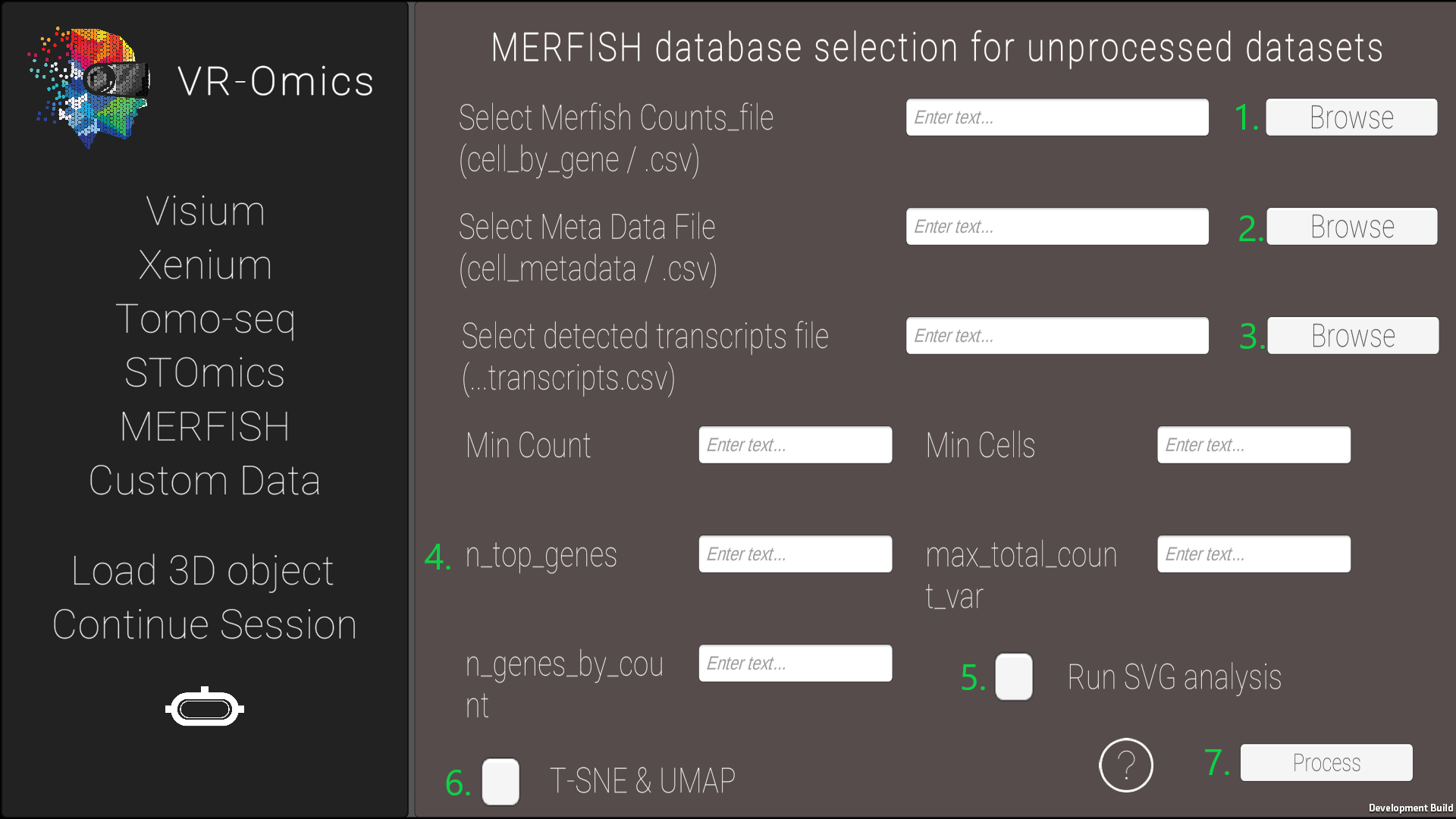
The MERFISH data we used is from Vizgen, where there data is publicly available under their data release program. 
p-value threshold for SVG cutoff can be customized. To modify the p-value, go The processed data will be saved in the VR-Omics data folder. To Visualise the data select the complete folder as shown in the figure 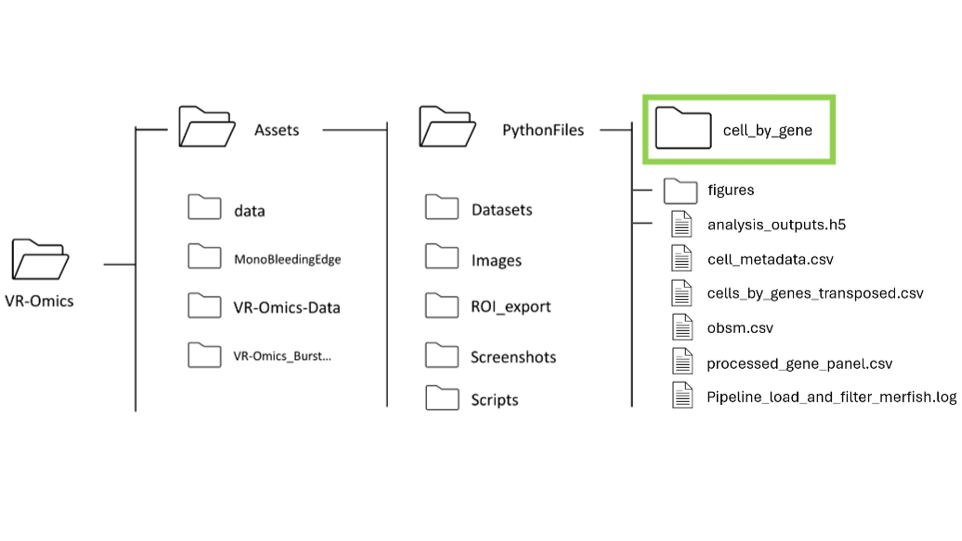
After clicking Process, the Python executable will be started. Don't close the pop up windows and wait until the pipeline has finished. This might take a couple of minutes. |
Load
| 1 | Navigate to the MERFISH load menu |
|

|
|---|---|---|---|
| 2 | Select the MERFISH files |
|
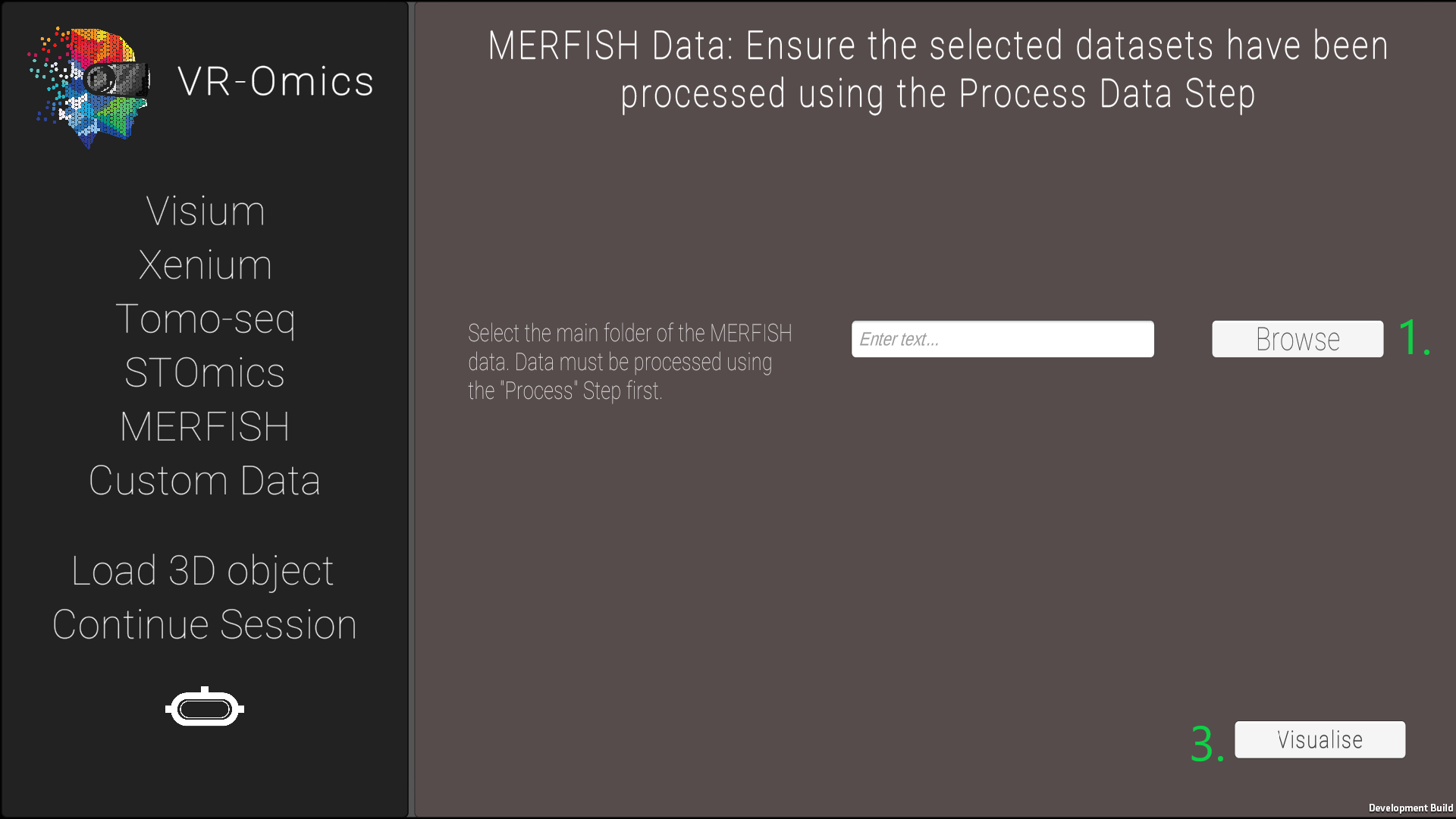
The processed data can be found in the VR-Omics data folder. To Visualise the data select the complete folder as shown in the figure 
The MERFISH data used was provided by Vizgen where there data is publicly available under their data release program. |
Merge 3D
| 1 | Navigate to the MERFISH Merge 3D menu |
|
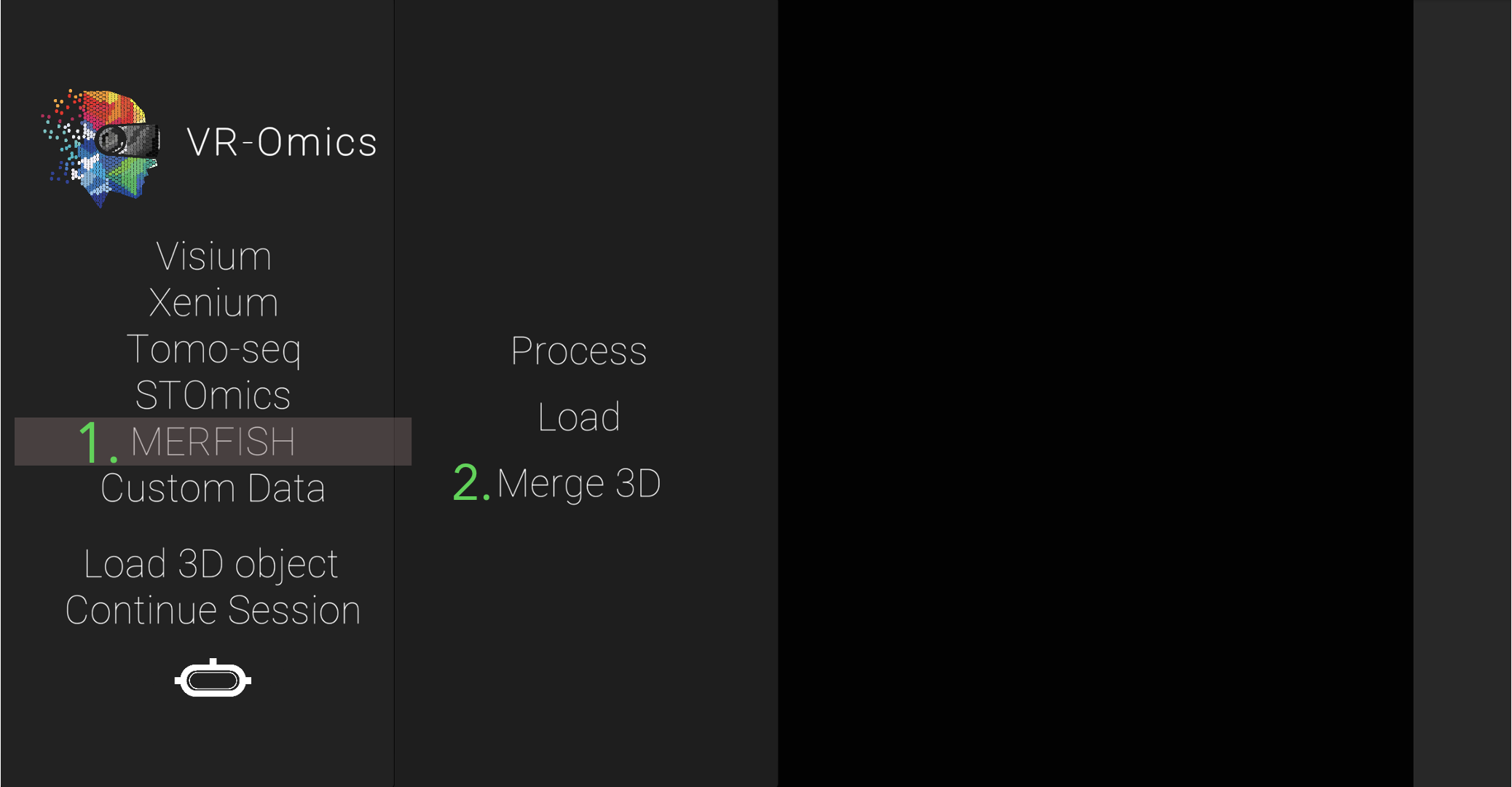
|
|---|---|---|---|
| 2 | Select sample folders |
|


../VR-Omics/Assets/PythonFiles/sample/ |
| 3 | Confirm selection |
|

If the tissue image wasn't found a default VR-Omics placeholder will appear. Please ignore if the respective file is named |
| 4 | Rotate the samples in the 3D mapping tool |
|
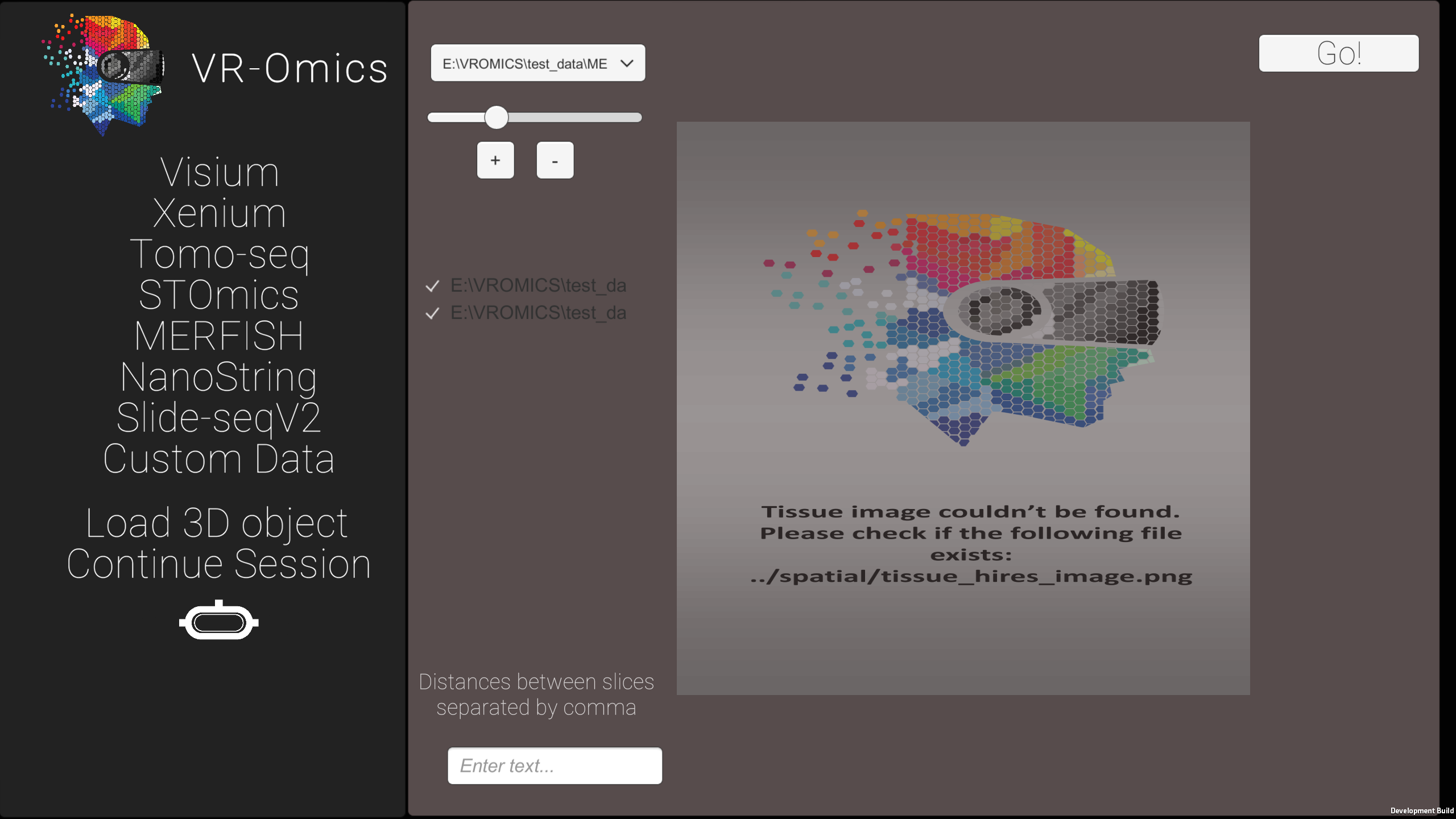
|
| 5 | Load the 3D dataset |
|


../VR-Omics/Assets/PythonFiles/sample/ |
NanoString
Process
| 1 | Navigate to the NanoString process menu |
|

|
|---|---|---|---|
| 2 | Select the NanoString files |
|

Selecting SVG Analysis and/or T-SNE/UMAP will increase processing time of the sample. 
p-value threshold for SVG cutoff can be customized. To modify the p-value, go The samples processed using the VR-Omics workflow are stored in the VR-Omics application folder on your local machine. 
After clicking Process, the Python executable will be started. Don't close the pop up windows and wait until the pipeline has finished. This might take a couple of minutes. |
Load
| 1 | Navigate to the NanoString load menu |
|

|
|---|---|---|---|
| 2 | Select the NanoString sample folder |
|
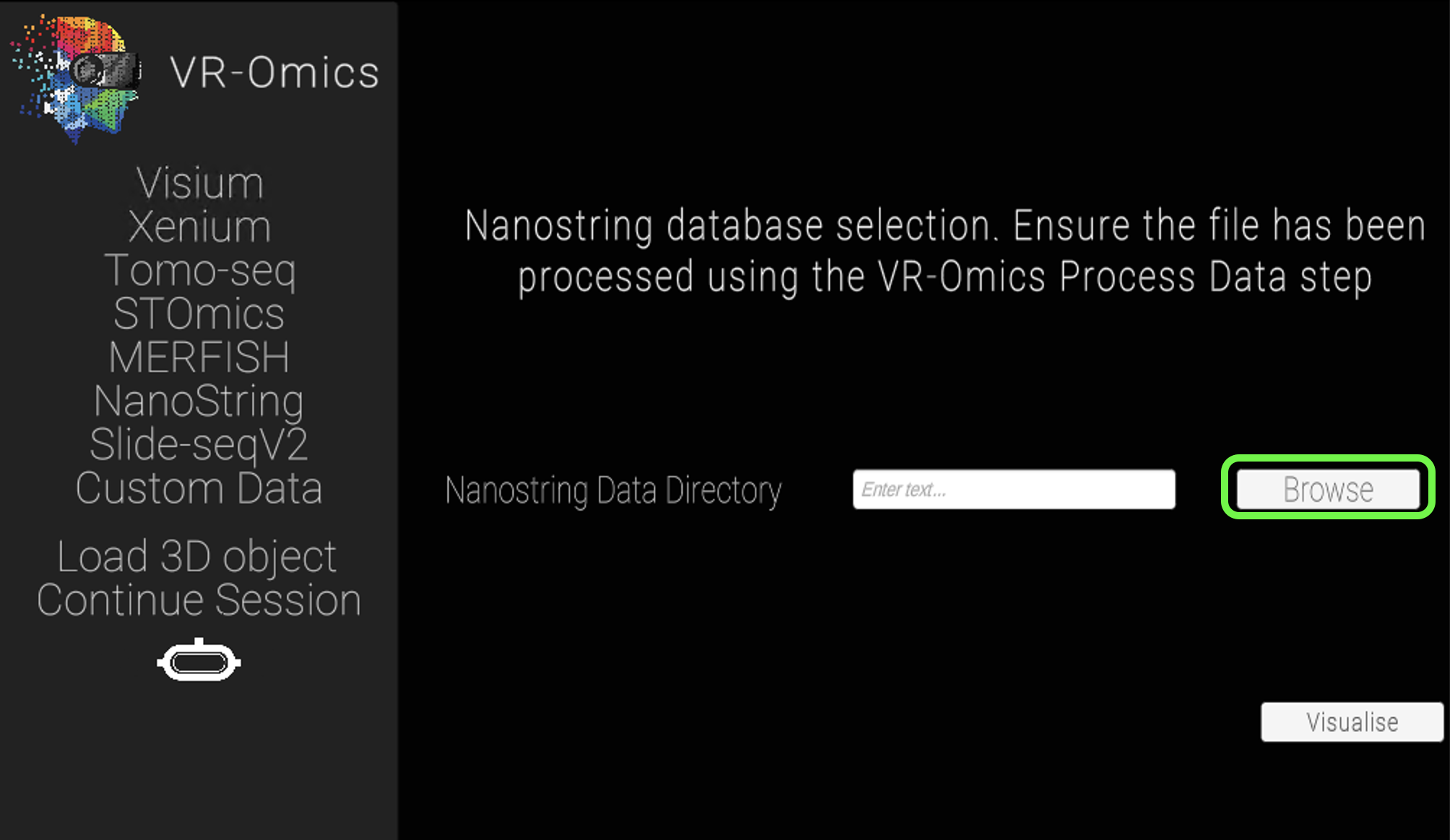
Select the NanoString folder processed with the VR-Omics AW (highlighted in green in the picture) using Browse. The processed data set can be found in the VR-Omics directory. Navigate from there to 
|
| 3 | Confirm selection |
Click Visualise to continue to the VR-Omics Visualiser.
|
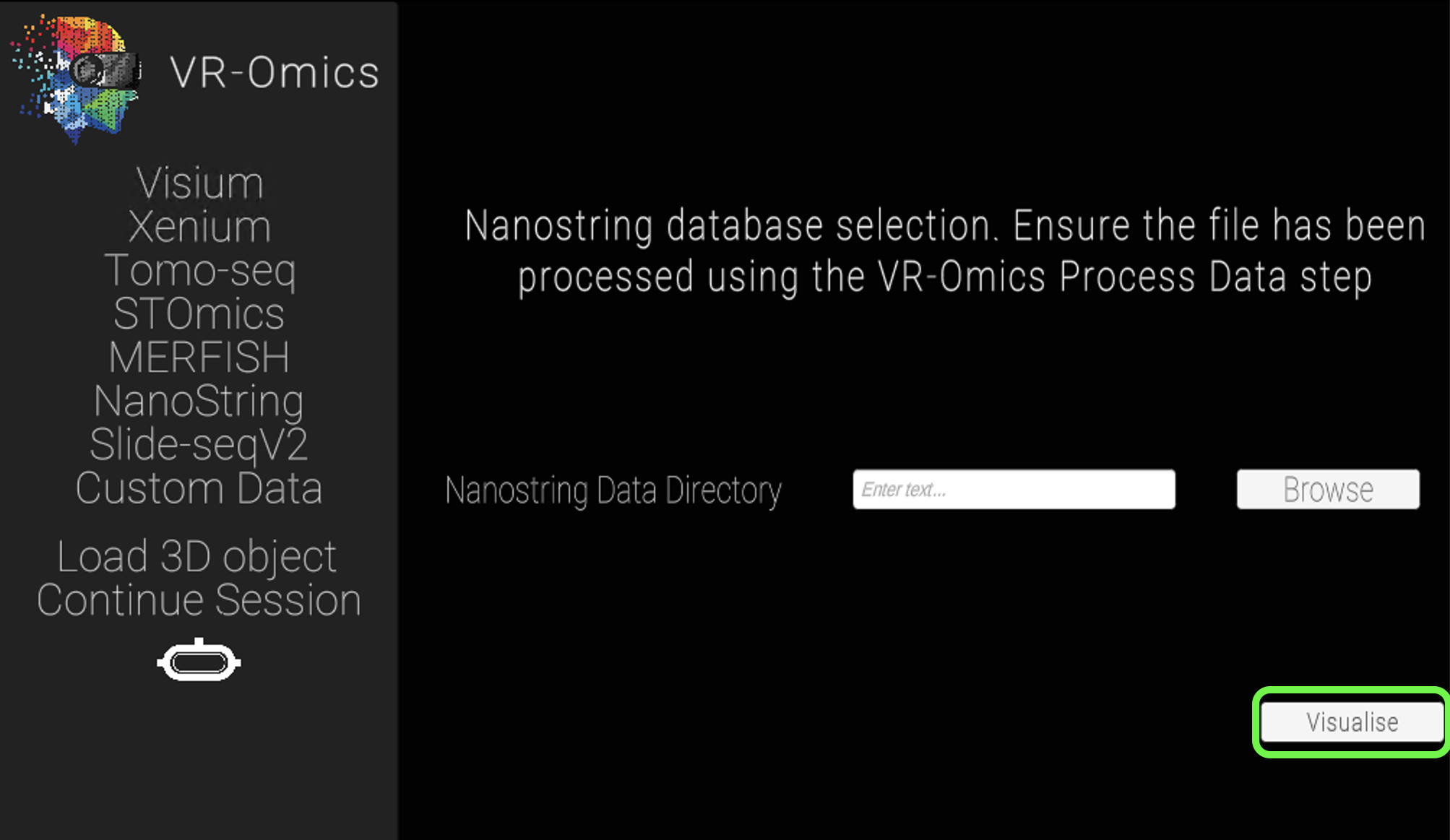
Ensure the data has been processed before visualising it using the Process function. |
Slide-SeqV2
Load
| 1 | Navigate to the Slide-SeqV2 load menu |
|

|
|---|---|---|---|
| 2 | Select the Slide-SeqV2 folder |
|
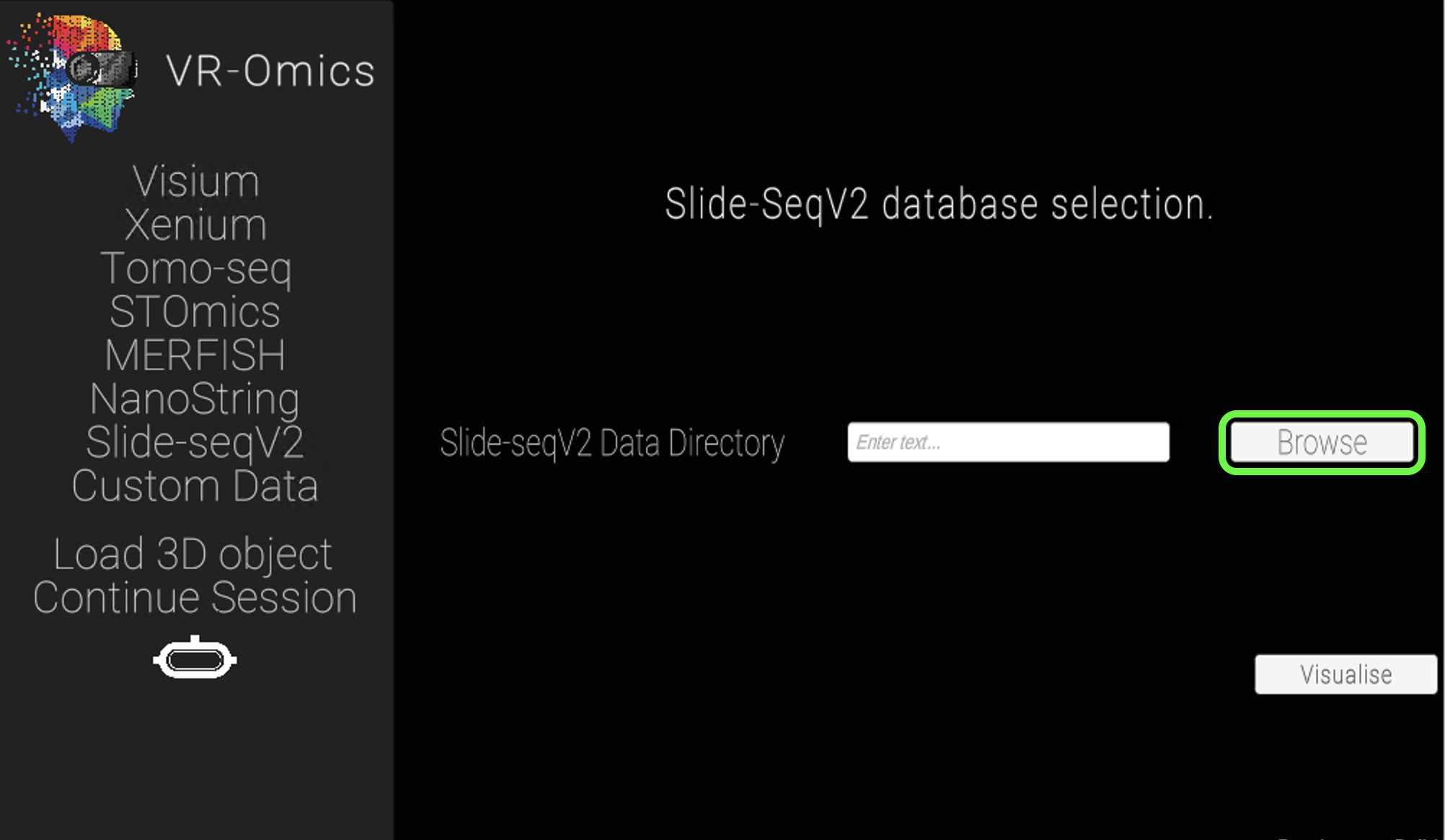
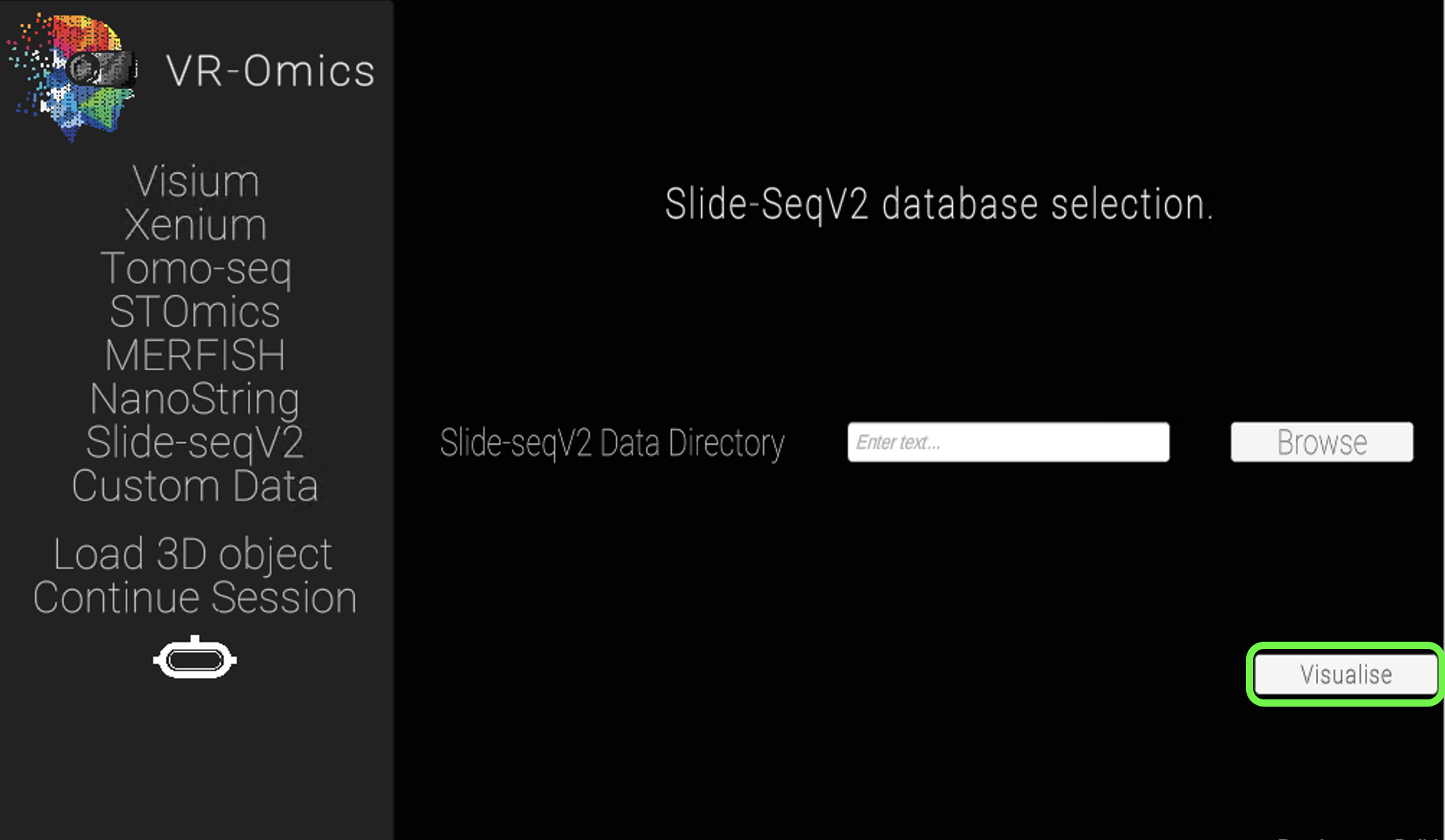
|
Required Folder Format for samples processed with Slide-SeqV2
For samples processed with Slide-SeqV2 workflow, please ensure this folder structure. Further descriptions of the files can be found below.

| Required file name ending | Description |
|---|---|
CellComposite
|
A folder consists cell composite .jpg images.
|
CellLabels
|
A folder consists cell labelling .tif images.
|
CellOverlay
|
A folder consists cell overlaying .jpg images.
|
CompartmentLabels
|
A folder consists cell compartment labelling .tif images.
|
exprMAt_file.csv
|
Header: in_tissue, array_row, array_col, n_genes_by_counts, log1p_n_genes_by_counts, total_counts, log1p_total_counts, pct_counts_in_top_50_genes, pct_counts_in_top_100_genes, pct_counts_in_top_200_genes, pct_counts_in_top_500_genes, total_counts_mt, log1p_total_counts_mt, pct_counts_mt, n_counts, n_genes, clusters
|
fov_positions_file.csv
|
CSV file with x and y pixel positions for all genes in the dataset.
|
metadata_file.csv
|
CSV file with one column header name gene listing all gene names in the dataset.
|
tx_file.csv
|
CSV file includes gene ID, length, and other related spatial transcript information about the data. |
Custom data
Load
| 1 | Navigate to the Custom data load menu |
|
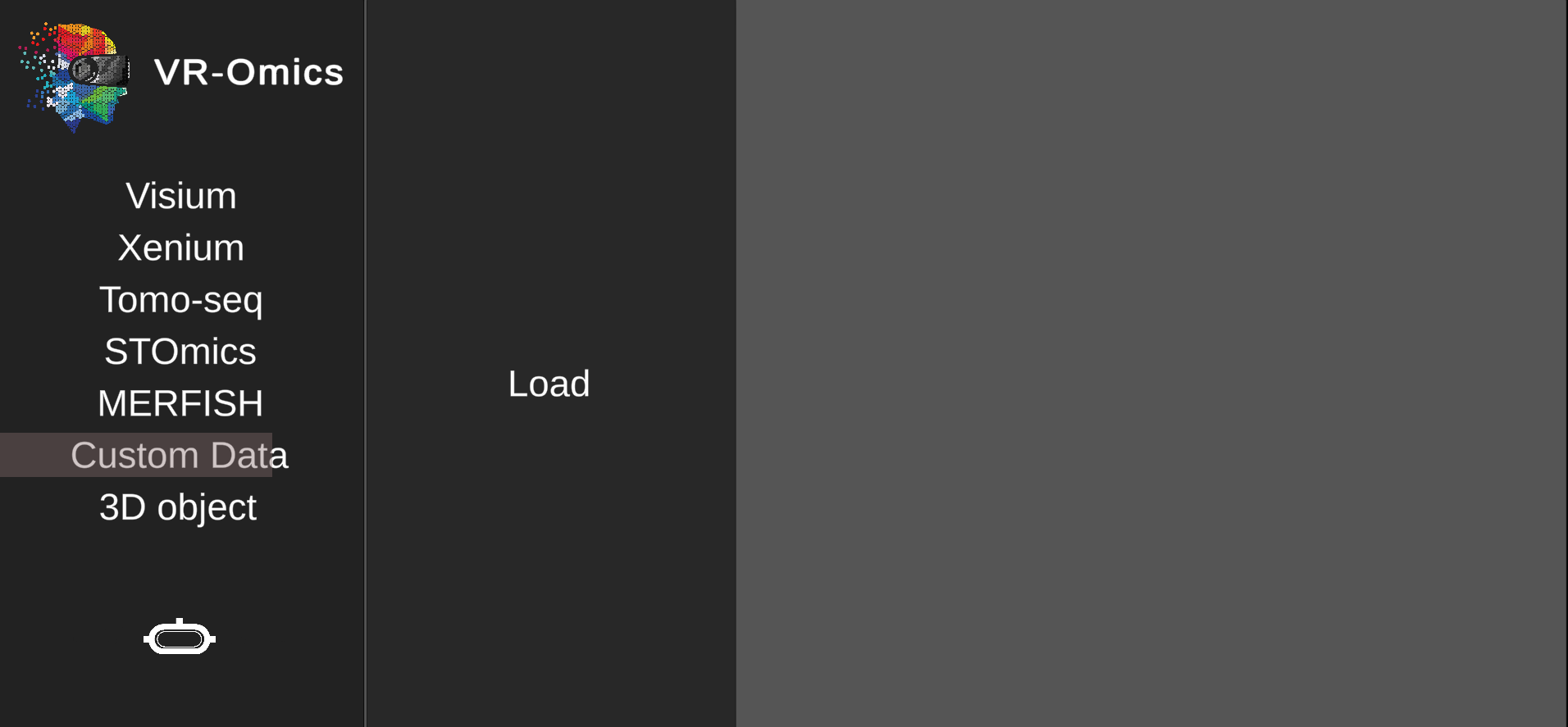
|
|---|---|---|---|
| 2 | Select the Custom data files |
.csv file using Browse.csv file using Browse |
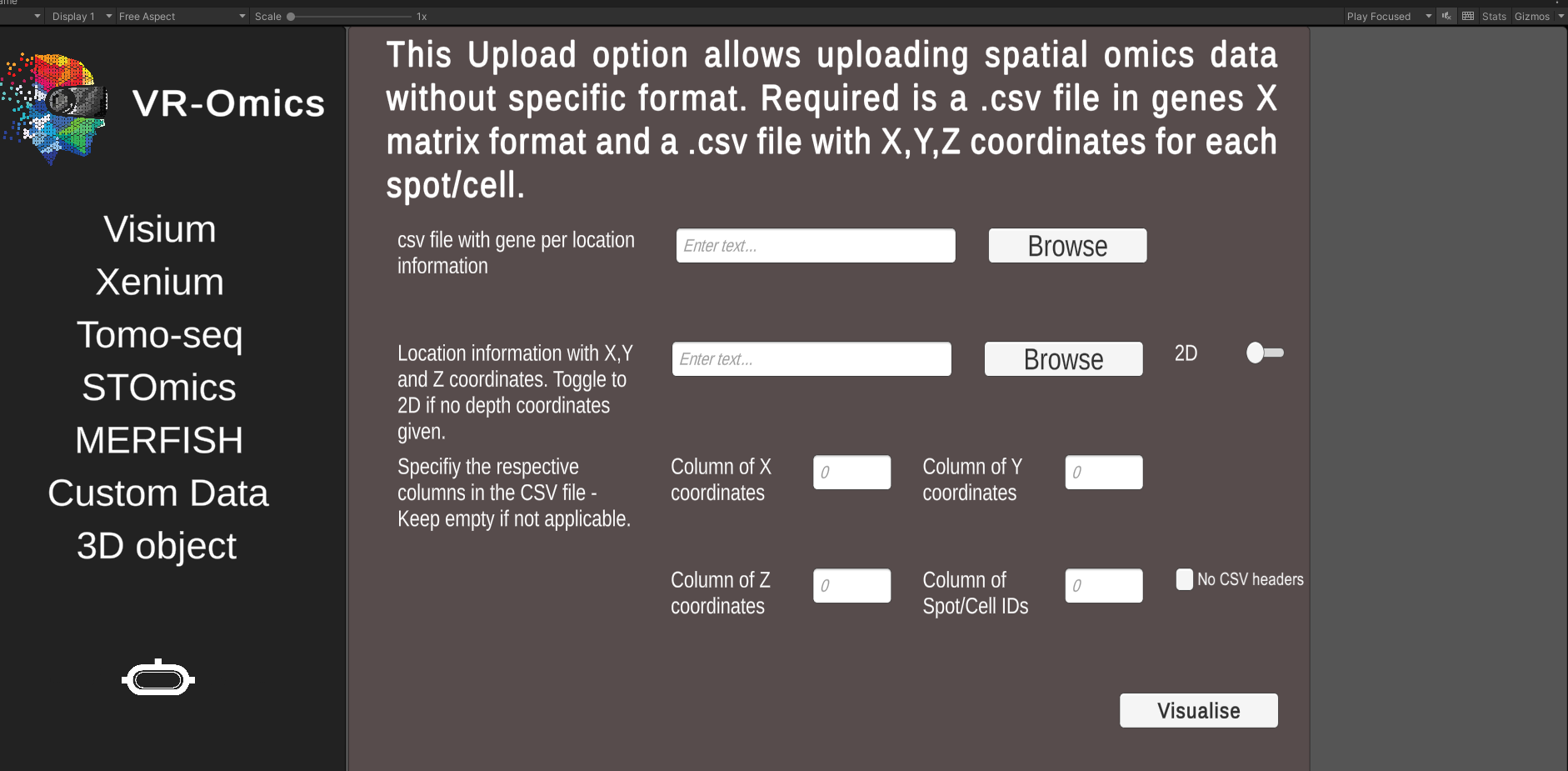
* Ensure the data is stored in a gene per location data file using columns for locations and rows for gene expressions ** The meta data file stores the X,Y and optional Z coordinates of the dataset. Each row needs to represent one location where the locations need to be in same order as in the gene expression file. |
| 3 | Specify columns |
0 for the second column enter 1 etc.0 for the second column enter 1 etc.0 for the second column enter 1 etc.*0 for the second column enter 1 etc.** |

* Leave blank if no three dimensional dataset ** Leave blank if no IDs available *** The header is the first row in a csv file usually used to store the headers of each column. |
3D Object
Load
| 1 | Navigate to the 3D object menu |
From the main menu select 3D object |
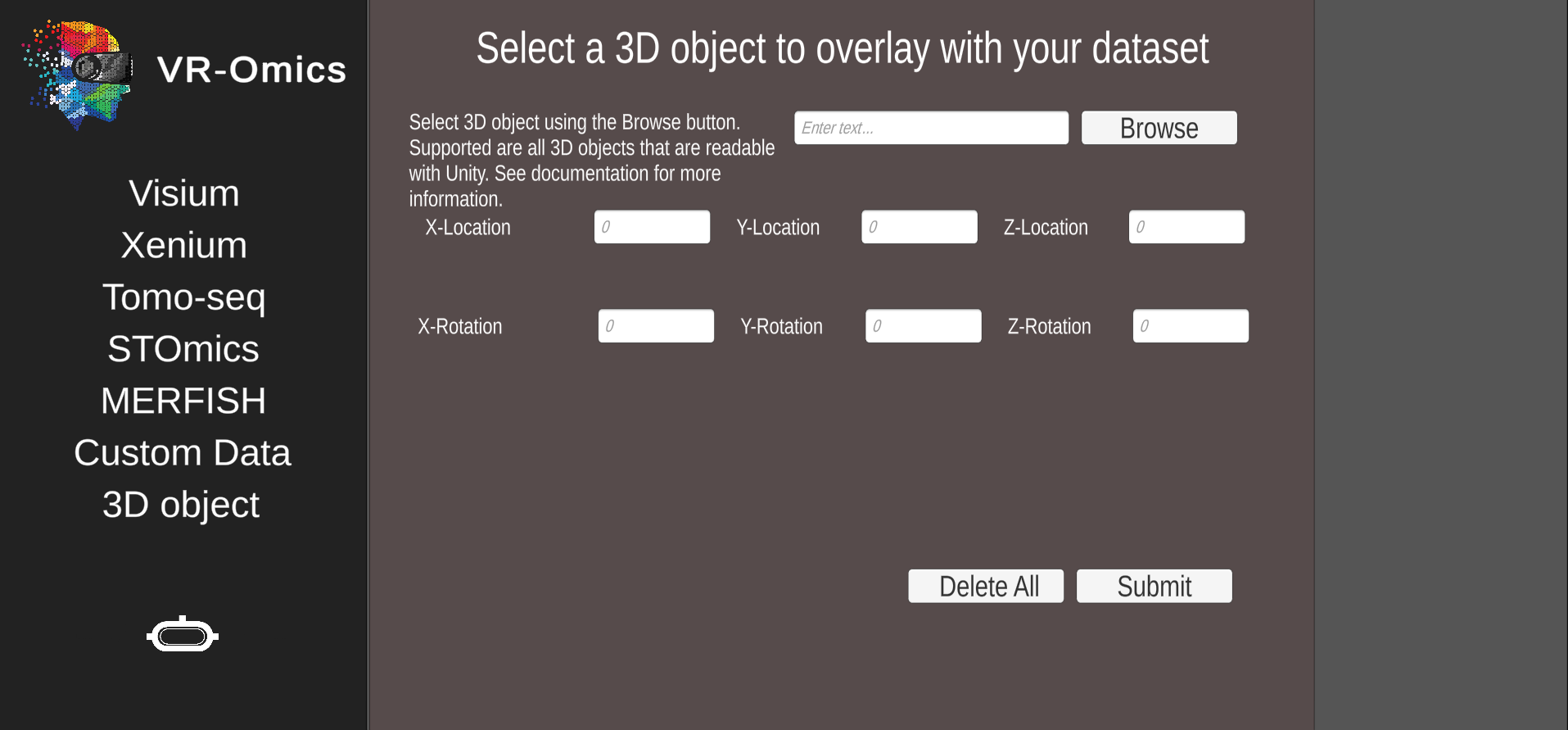
|
|---|---|---|---|
| 2 | Select the 3D object file |
|

* Only 3D object supported by Unity can be uploaded. More information can be found using the Unity documentation. This feature was tested with ** The coordinates are relative to the dataset, the dataset will always be rendered in the origin *** The rotation is relative to the dataset, the dataset will always be rendered in the origin Ensure the 3D object has the right measurements, the object can be resized within the VR-Omics Visualiser but if the object was uploaded top big, or too small compared to the rendered dataset this might lead to issues. In this case please adapt the dimensions of the object prior. |
| 3 | Submit the object |
Store the selected 3D object by clicking Submit |

Ensure the object was submitted before uploading a spatial dataset. This can't be done after entering the VR-Omics Visualiser. |
| 4 | Removing an object |
Remove all selected objects from the selection using Delete All |

Multiple objects can be uploaded, although this feature is only tested with one object. |
AW Packages used
| Name of package | Description | Platforms used | Link |
|---|---|---|---|
| Scanpy |
Scanpy is an open-source toolkit for the analysis of single-cell and spatial expression data. |
|
Scanpy |
| Squidpy |
Squidpy is a tool for the analysis and visualization of spatial molecular data. |
|
Squidpy |
| Stereopy |
Stereopy is a package for spatial transcriptomics data analysis of STOmics data. |
|
Stereopy |
| Matplotlib |
Matplotlib is a Python 2D plotting library. |
|
Matplotlib |
| Seaborn |
Seaborn is a data visualization library based on Matplotlib. |
|
Seaborn |
| SpatialDE |
SpatialDE is a method to identify genes which significantly depend on spatial coordinates in non-linear and non-parametric ways. |
|
SpatialDE |
| LeidenAlg |
LeidenAlg is a Python implementation of the Leiden community detection algorithm. |
|
LeidenAlg |
| Pandas |
Pandas is a data manipulation and analysis library for Python. |
|
Pandas |
| NumPy |
NumPy is a fundamental package for scientific computing with Python. |
|
NumPy |
| Scanorama |
Scanorama enables batch-correction and integration of heterogeneous scRNA-seq datasets. |
|
Scanorama |
VR-Omics Visualisation
Gene Search Features
| Feature | Description | Icon | Image |
|---|---|---|---|
| Gene search - Heatmap
All VR Demo |
To search for a gene (heatmap) using the search bar:
|
|
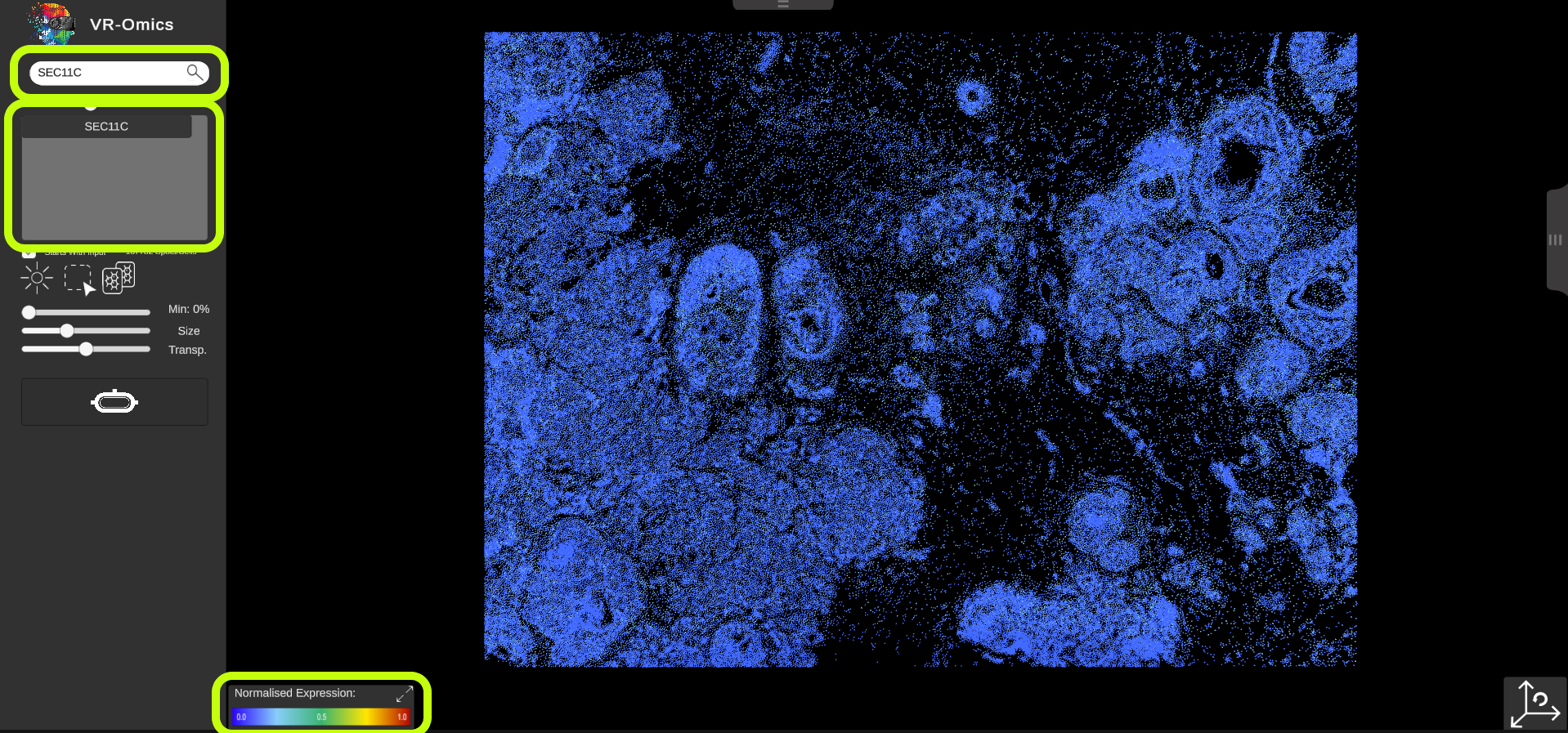
|
| Binary search - Gene is on
Visium Xenium STOmics Merfish Custom VR Demo |
To search for a gene (binary) using the search bar:
|
|
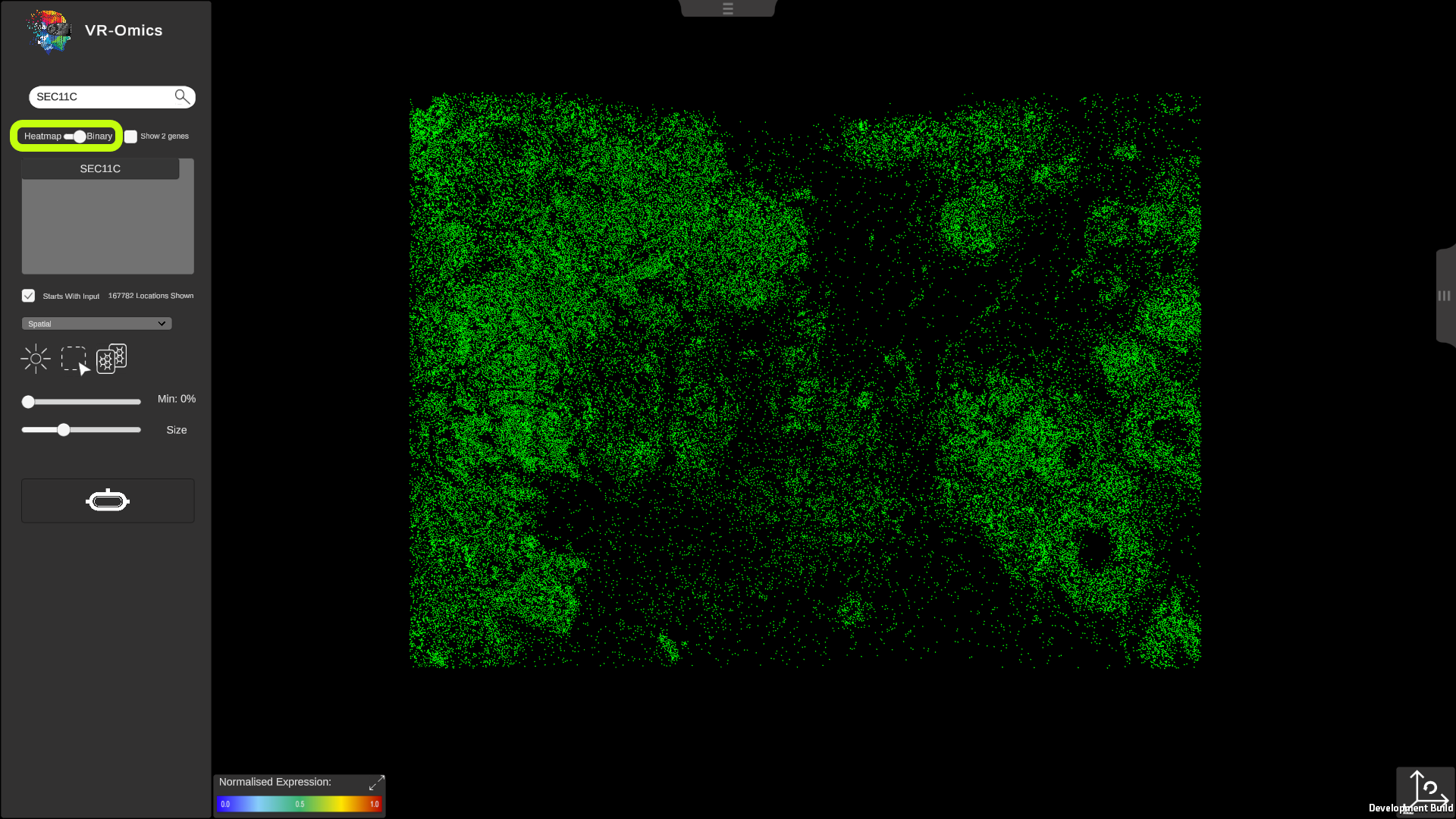
|
| Change if gene search contains or starts with term
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
By default, gene names entered into the search bar will only return results that start with the input. To also include gene names that contain the input, uncheck the
|
|
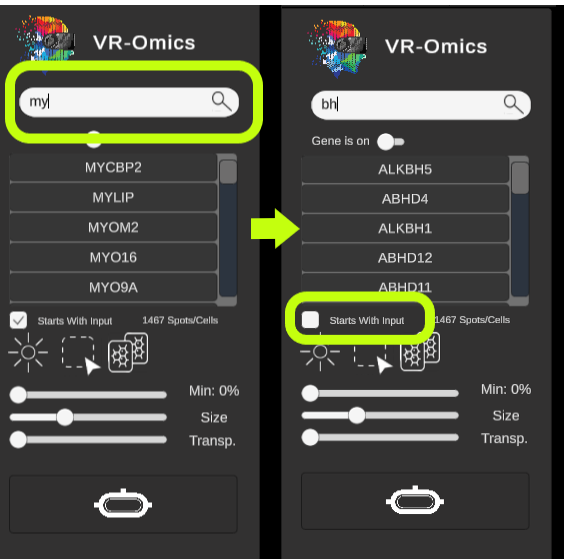
|
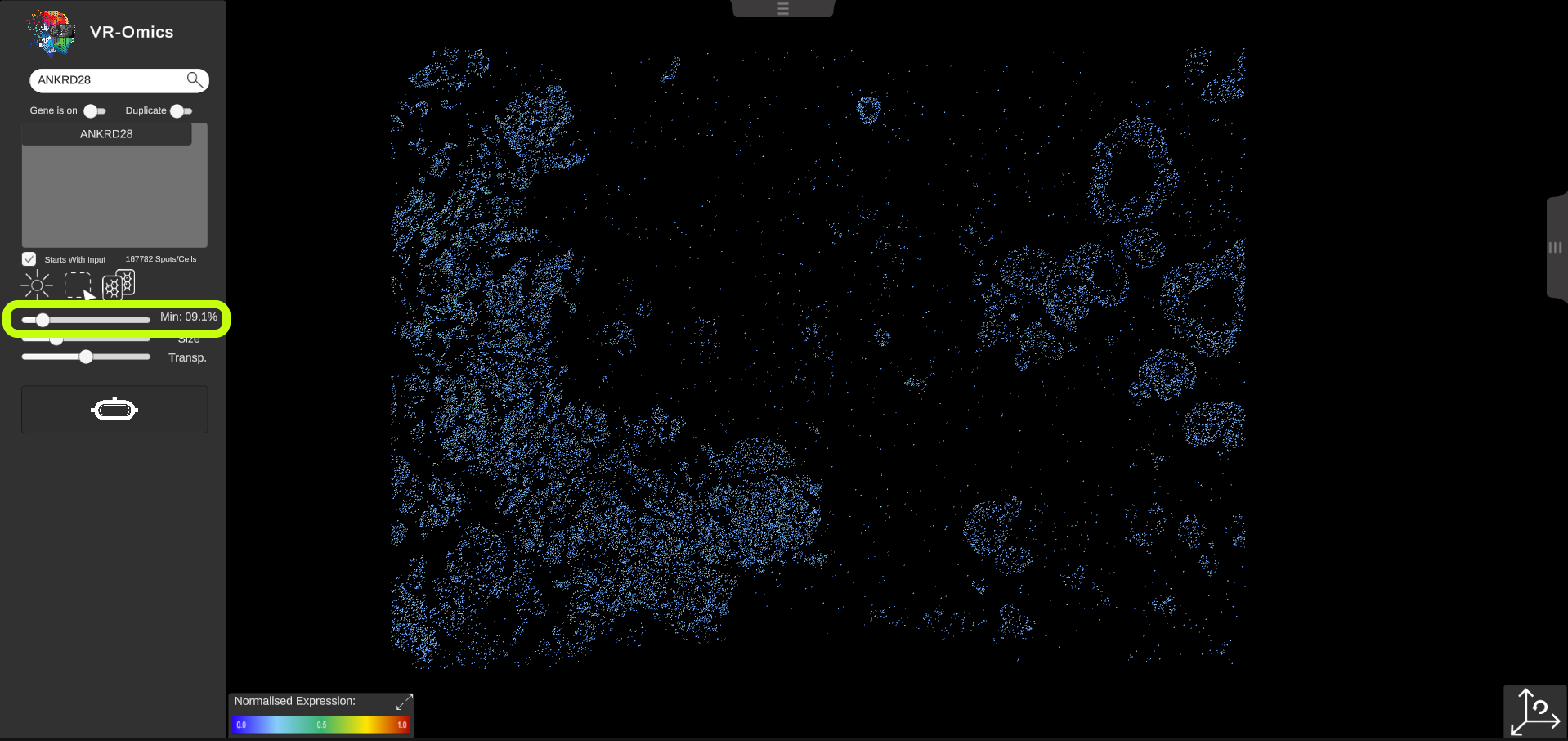
| Threshold - Minimum
Visium Xenium STOmics Tomo-Seq Merfish Custom VR Demo |
By default, each location in the dataset is coloured according to the normalised gene expression values. If no value is found for a location, the location will be cleared. The threshold slider is set to 0% by default, which means that all locations with an expression value of 0 or Lower of the normalised values will be visualised. To focus on regions with a Low expression level of the gene, you can adjust the slider to a Lower position:
|
|
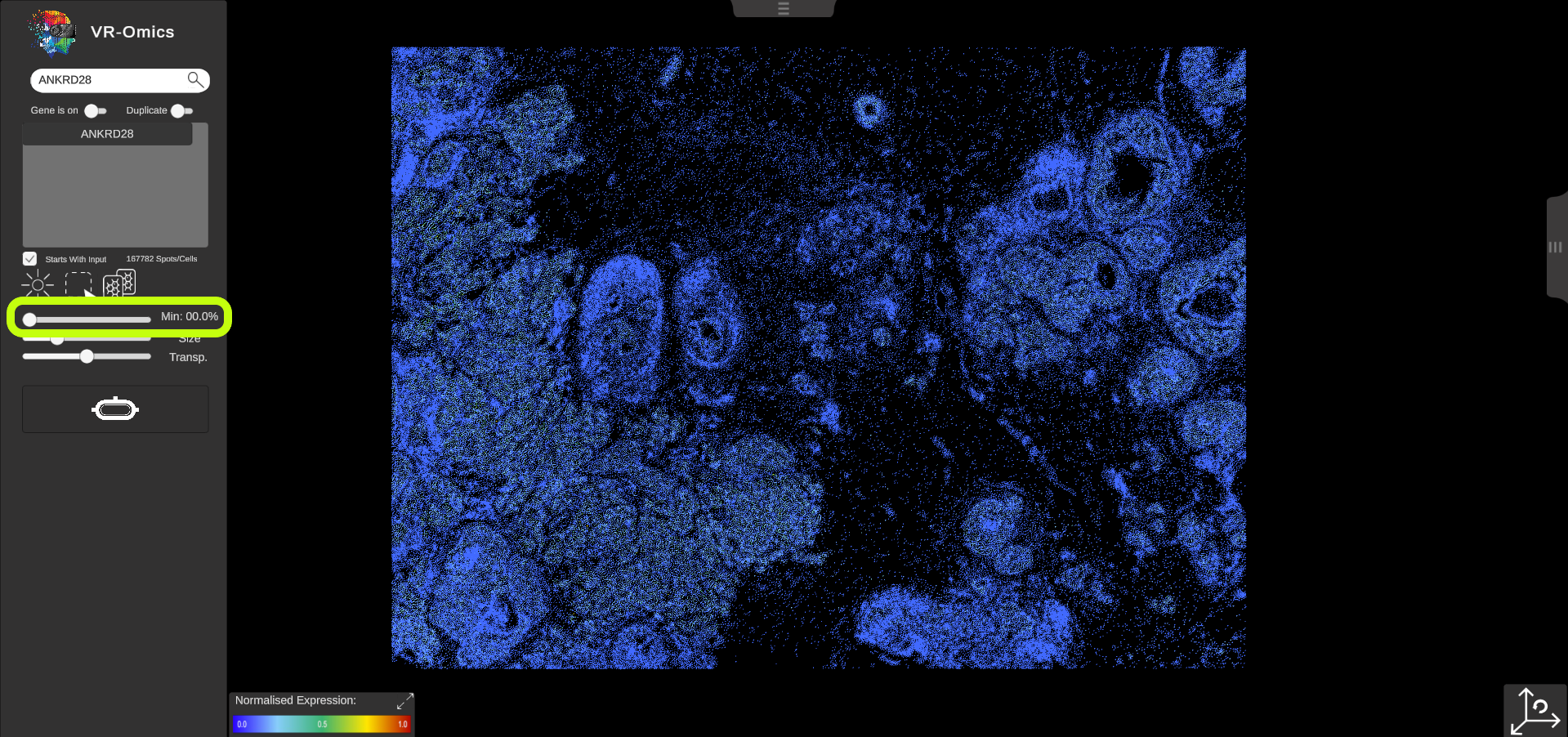
|
Location Interaction Features
| Feature | Description | Icon | Image |
|---|---|---|---|
| Selecting Regions of Interest (ROI)
Visium Xenium STOmics Merfish Custom VR Demo |
ROIs can be selected by using the selection tool. To select a group of ROIs:
|
|
|
| Remove a single location
Visium Xenium STOmics Merfish Custom VR Demo |
Single locations can be removed from the ROI selection. To remove a single location from the ROIs:
|
|
|
| Remove all selections
Visium Xenium STOmics Merfish Custom VR Demo |
All locations can be removed from all groups at once. To remove all selections:
|
|
|
| Export ROI selections
Visium Xenium STOmics Merfish Custom VR |
ROI selections can be exported. This will export all current selected locations and the respective groups they are in for later analysis. To export selections:
The export function will override the last selection. Save the selection locally by copying the export file.
|
|
|
| Import ROI selections
Visium VR |
To import previously saved ROI selections:
The export function does not check if the same dataset is used. Importing the last selection into a different dataset will cause errors. Ensure using the same data set to import as the one the export was generated from.
|
|
|
| 3D ROI selection
Visium Xenium STOmics Merfish Custom VR Demo |
In VR-Omics, it is possible to make 3D selections of ROIs if a 3D dataset (e.g. multiple Visium slides) is used. To make a 3D selection:
This allows the user to select 3D regions of interest. These selections can be exported as described above. To toggle between 3D and 2D selection, use the 3D selection button. |
|
|
Dimensionality Reduction Features
| Feature | Description | Icon | Image |
|---|---|---|
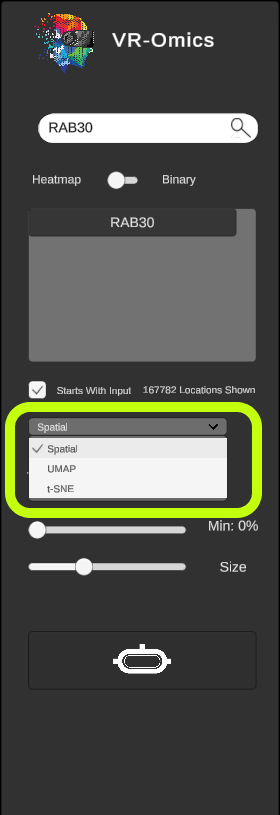
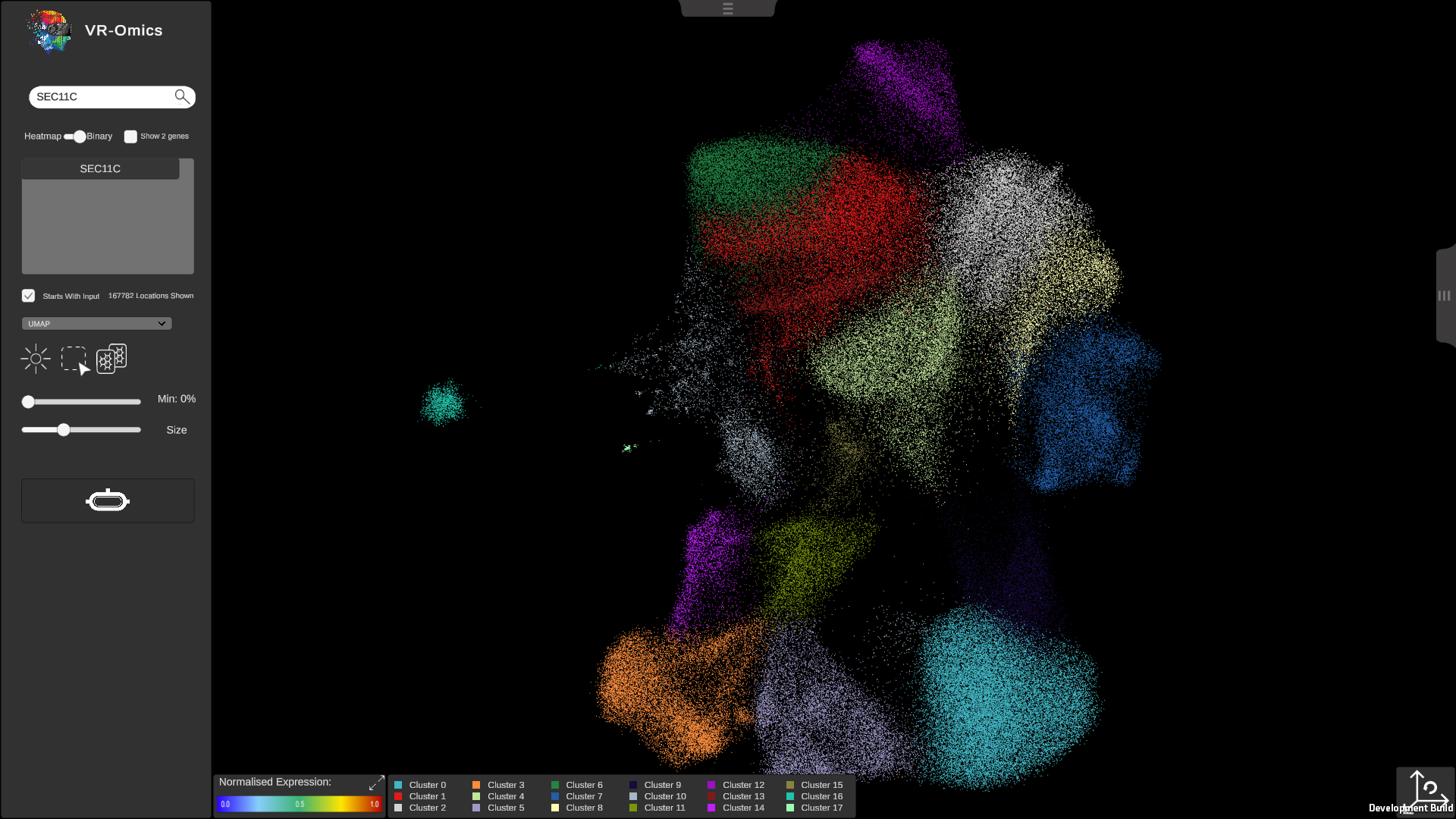
| Visualising UMAP
Visium Xenium STOmics Merfish VR |
|
|
| Visualising t-SNE
Visium Xenium STOmics Merfish VR |
|

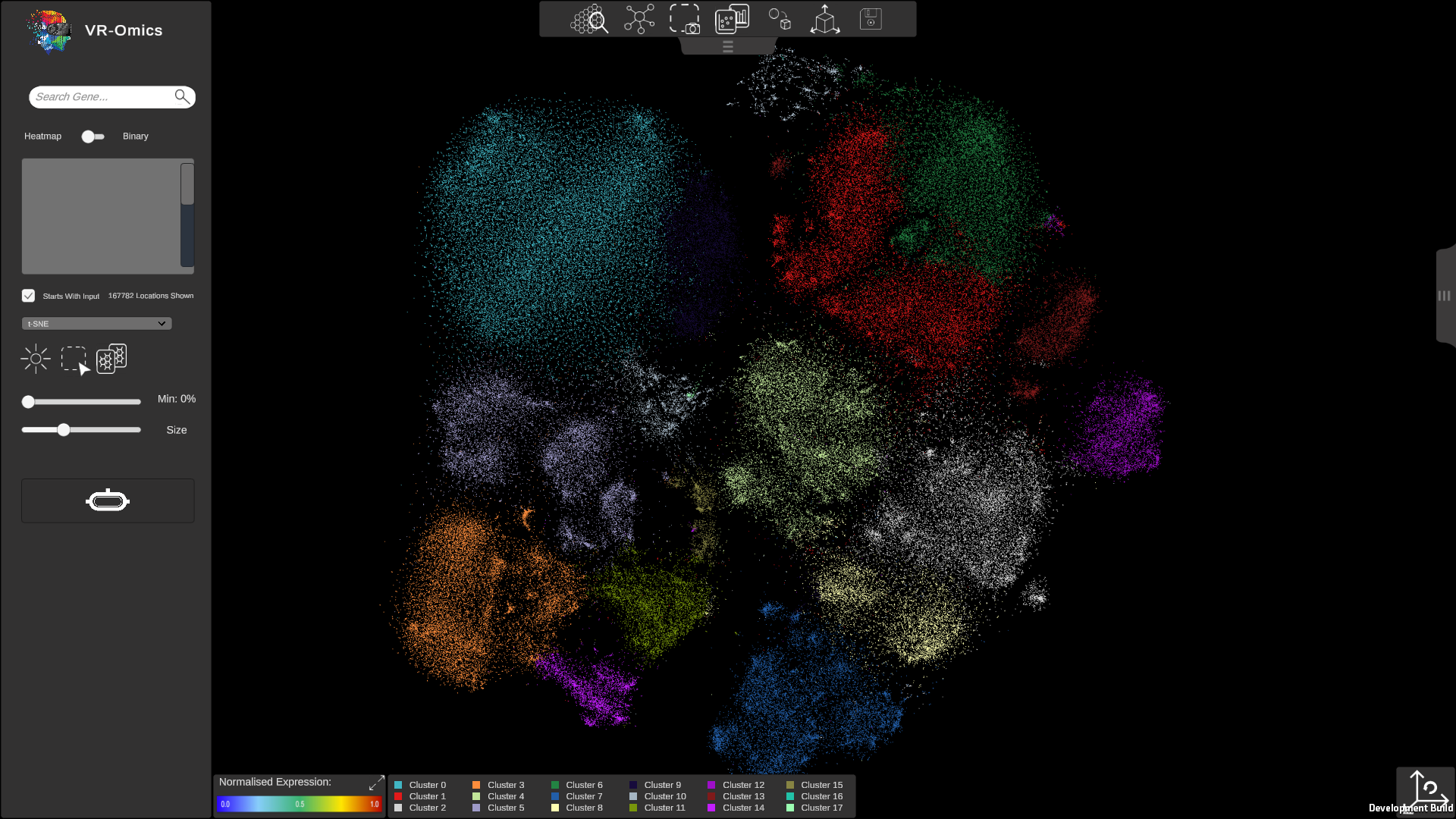
|
| Visualising spatial coordinates
Visium Xenium STOmics Merfish VR |
|

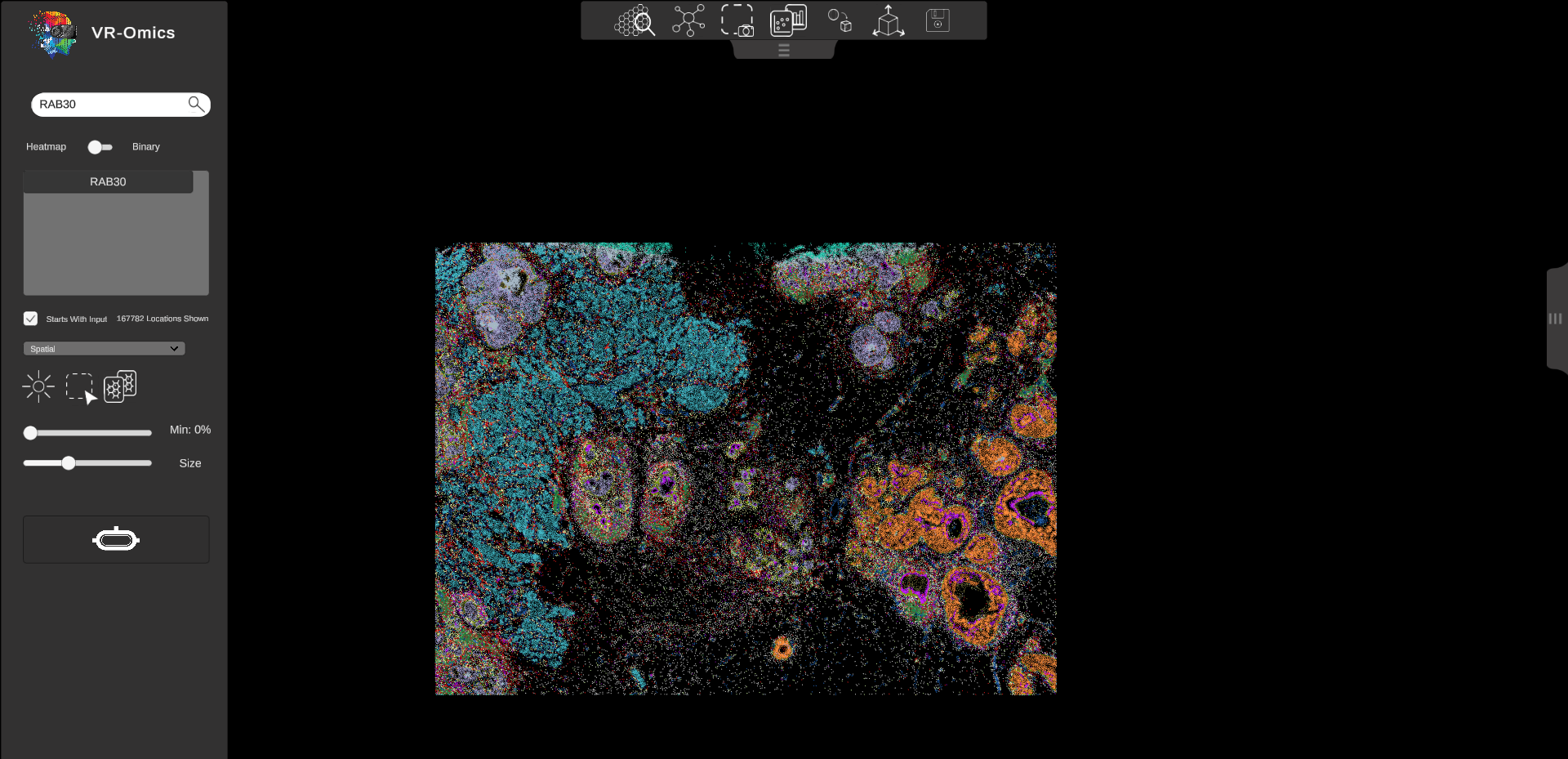
|
Cluster Visualisation Features
| Feature | Description | Icon | Image |
|---|---|---|---|
| Show Leiden Cluster
Visium Xenium STOmics Merfish Custom VR Demo |
If cluster information is available it can be visualised in the VR-Omics Visualiser:
|
|
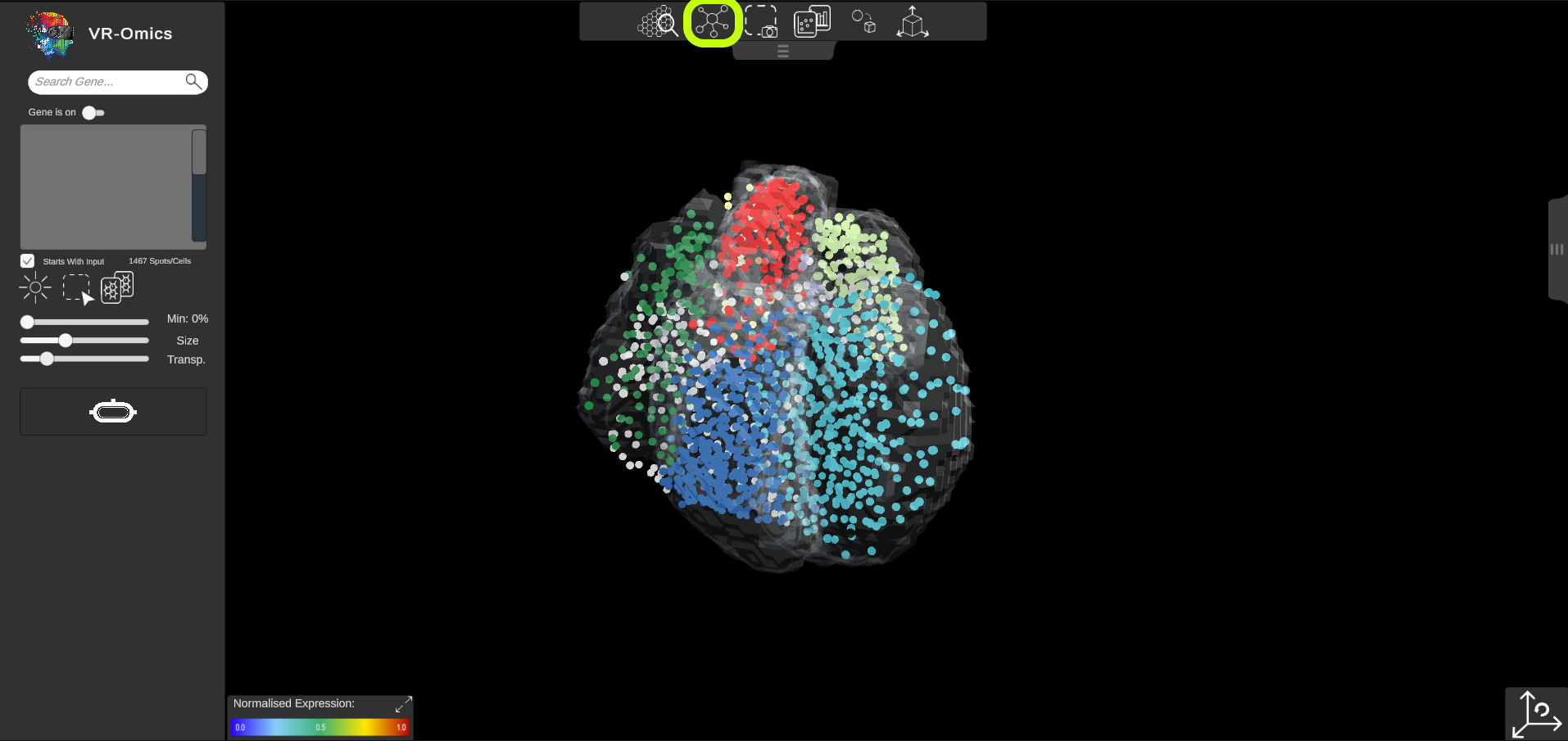
|
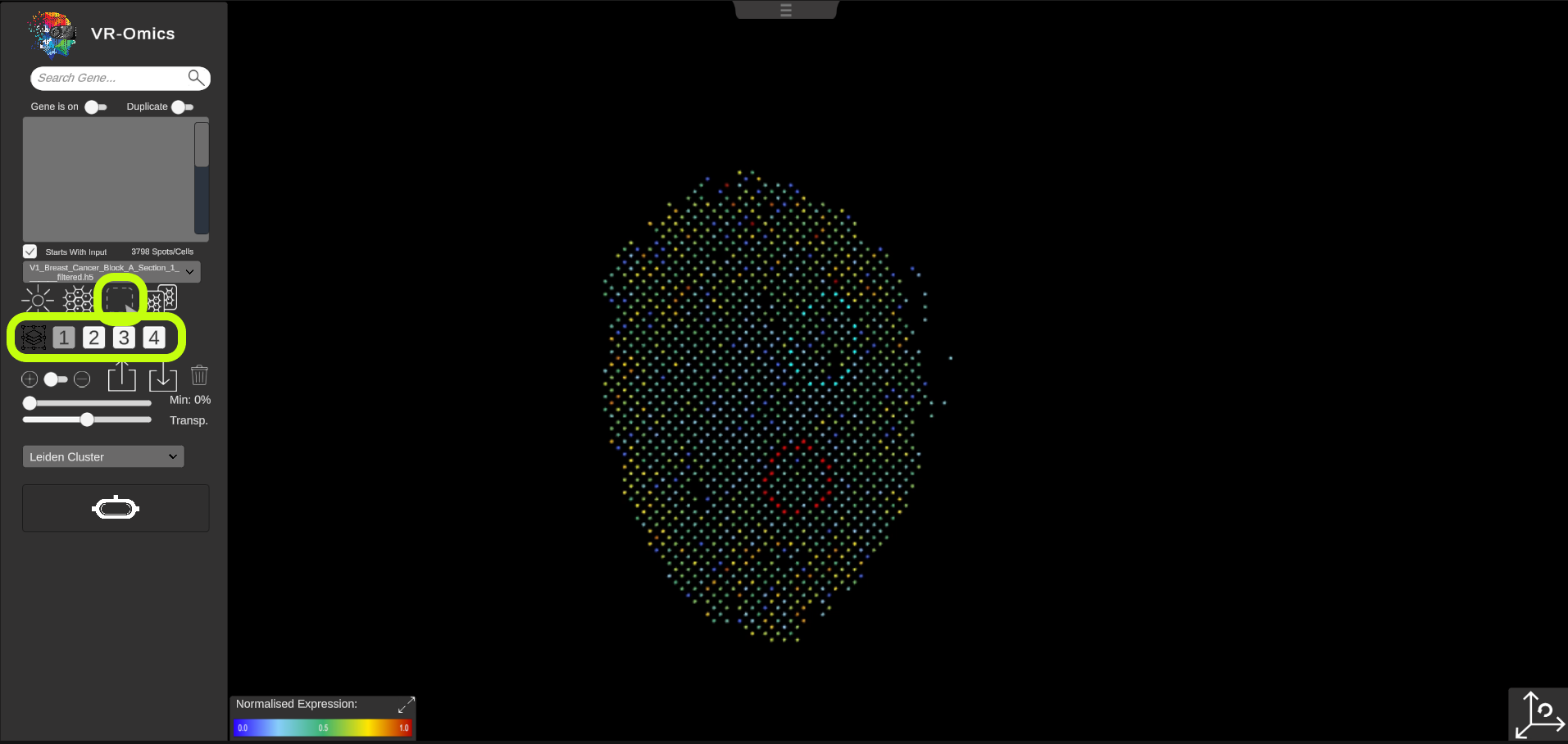
| Show Single Clusters
Visium Xenium STOmics Merfish Custom VR Demo |
|
|
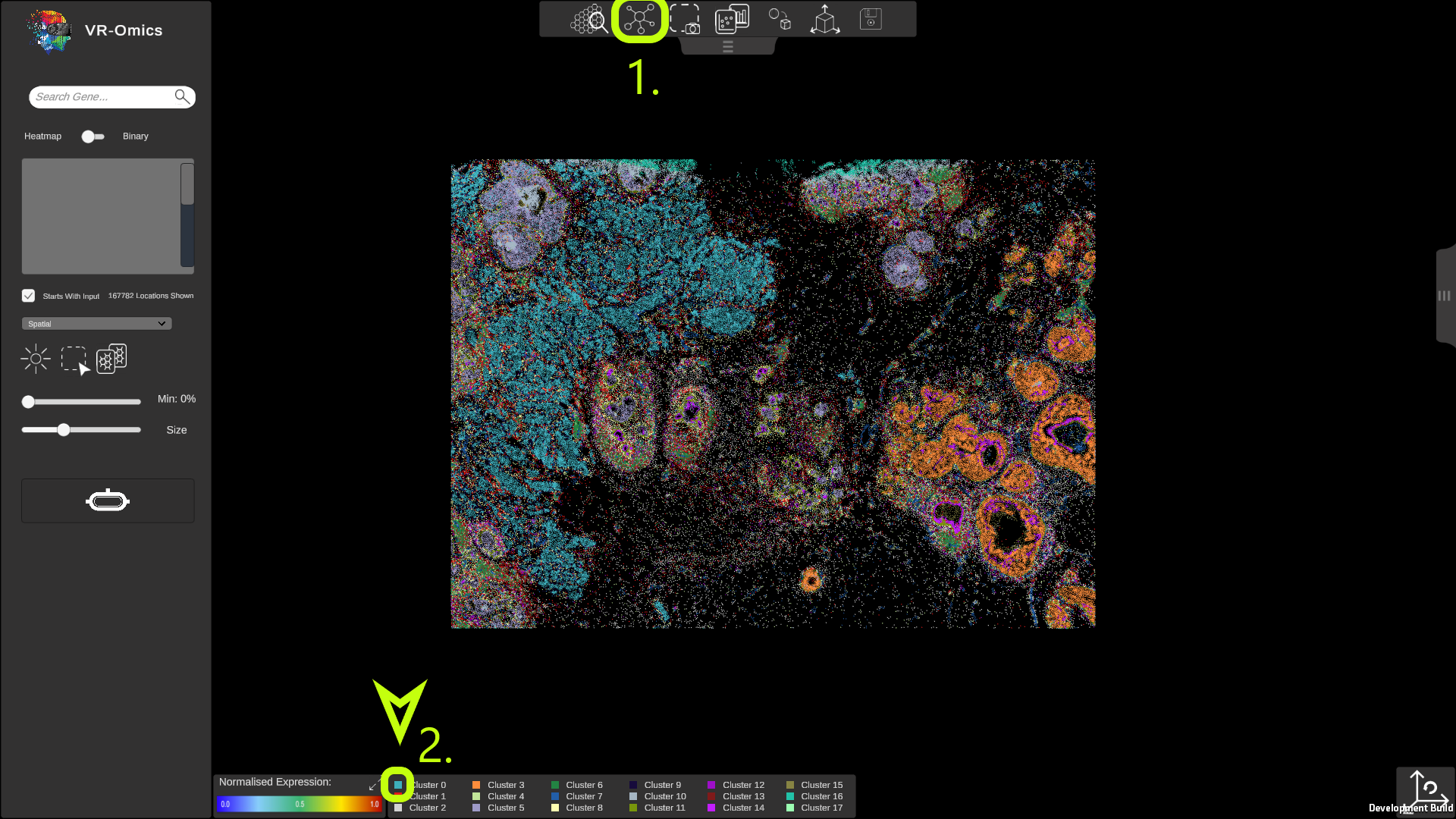
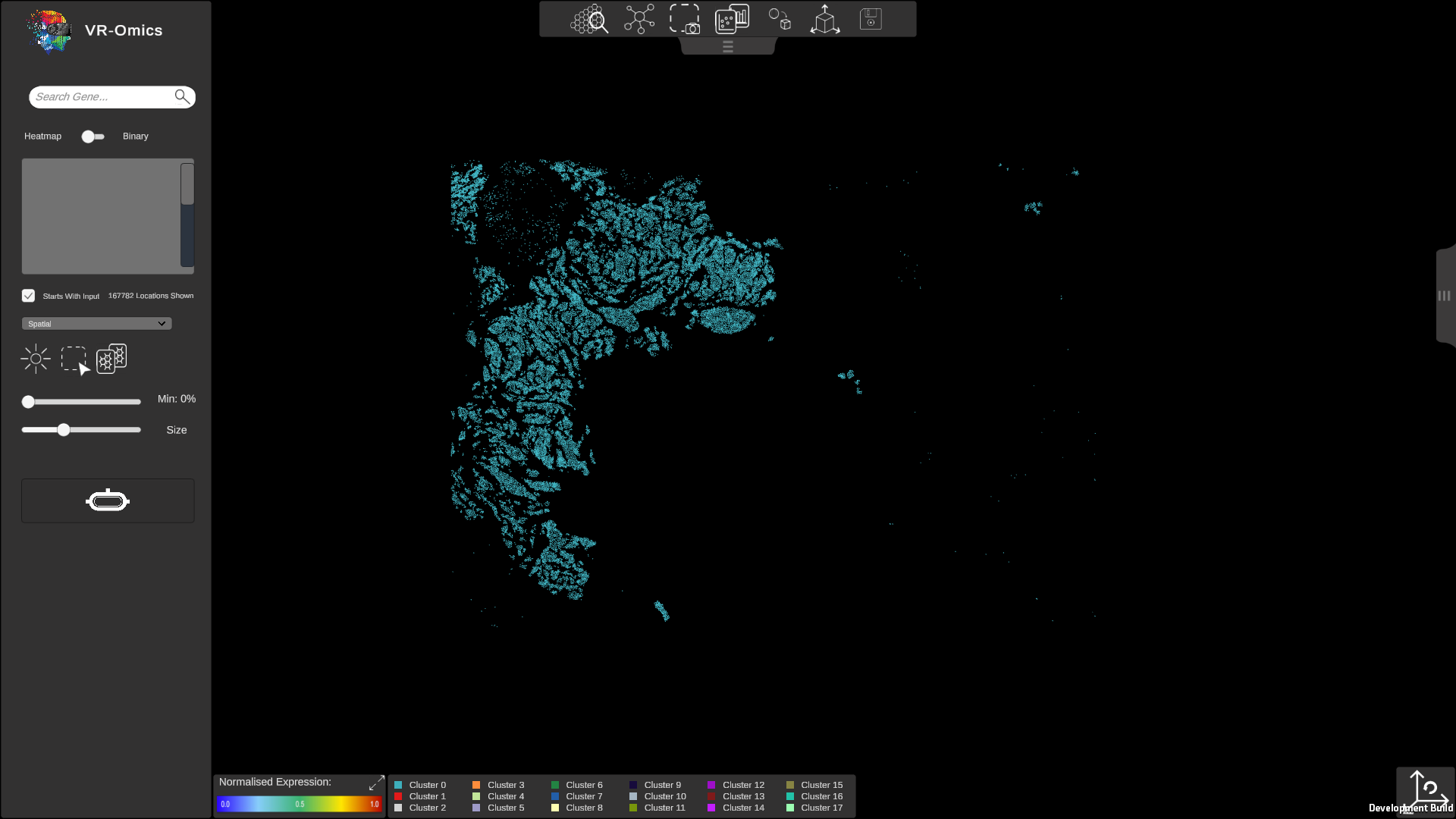
|
Tissue Overlay Features
| Feature | Description | Icon | Image |
|---|---|---|---|
| Tissue Overlay
Visium VR |
The dataset can be overlaid with the respective tissue image. Use the tissue button to activate the tissue image overlay. The image is aligned automatically. The image will be automatically downloaded if the dataset was downloaded using VR-Omics Automated Workflow. If using a different dataset, ensure that the tissue file is called The tissue image provides additional spatial orientation. |
|

More information about the tissue alignment can be found in the 10X Genomics Spatial Outputs section. |
| Tissue Opacity
Visium VR |
The tissue opacity can be adjusted using the opacity slider in the tissue context menu. Move the slider to the left decreased the tissue opacity. Move the slider to the right increases the tissue opacity. |
|
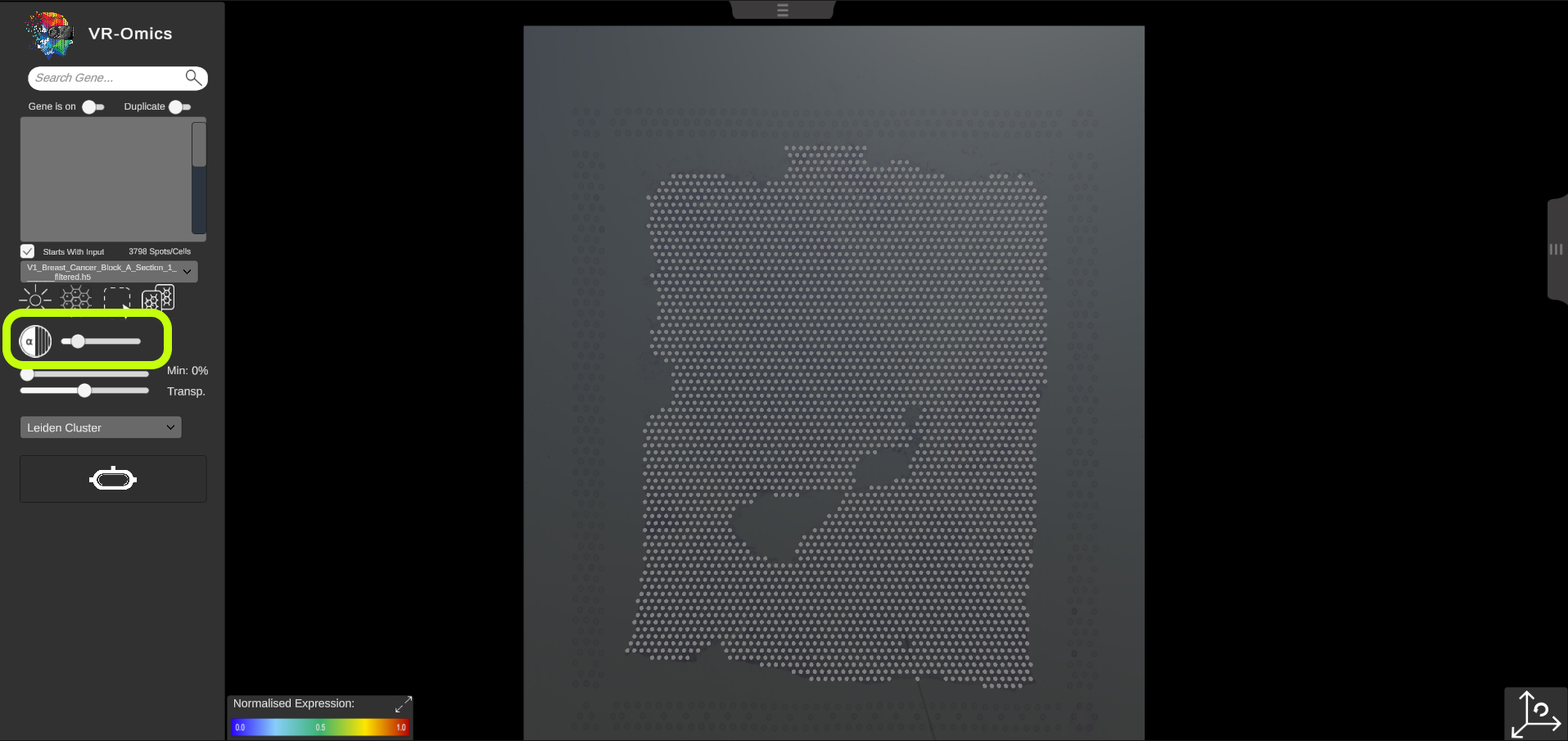

|
Explorative Comparison Features
| Feature | Description | Icon | Image |
|---|---|---|---|
| Side-by-Side comparison
Visium Xenium STOmics Merfish Custom VR Demo |
The side-by-side comparison feature allows comparing two different gene expression patterns next to each other. To enable the feature, click on the "Compare" button in the menu. The current dataset will be duplicated. Additionally, a slider with the label If the slider is set to left, and a new gene is selected from the search bar, the gene will be applied to the left dataset. If the slider is set to right, the next gene will be applied to the right side of the dataset. This feature can be combined with the ROI selection tool, where ROIs will be visualized in both datasets. |
|
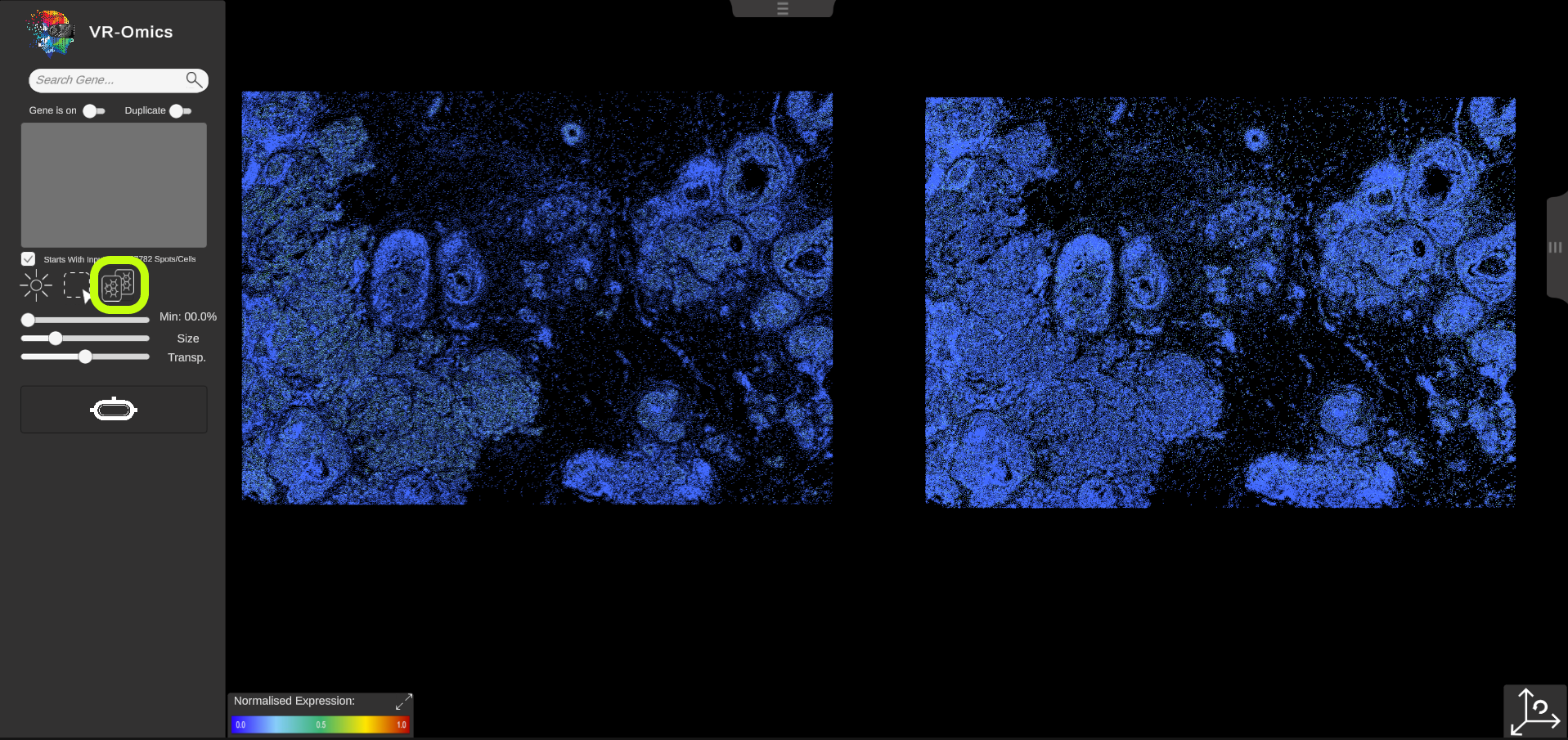
|
| Comparison of two gene expression patterns
Visium Xenium STOmics Merfish Custom VR Demo |
If the side-by-side comparison feature is disabled while two different genes were selected, a dialog menu will open asking to merge or discard the selection. If the user decides to merge the selection, a vector based difference analysis is performed. The difference between the both genes selected in each location will be calculated and visualised in a heatmap like manner. This allows easily to visualise regions of similarity or high differences. |
|
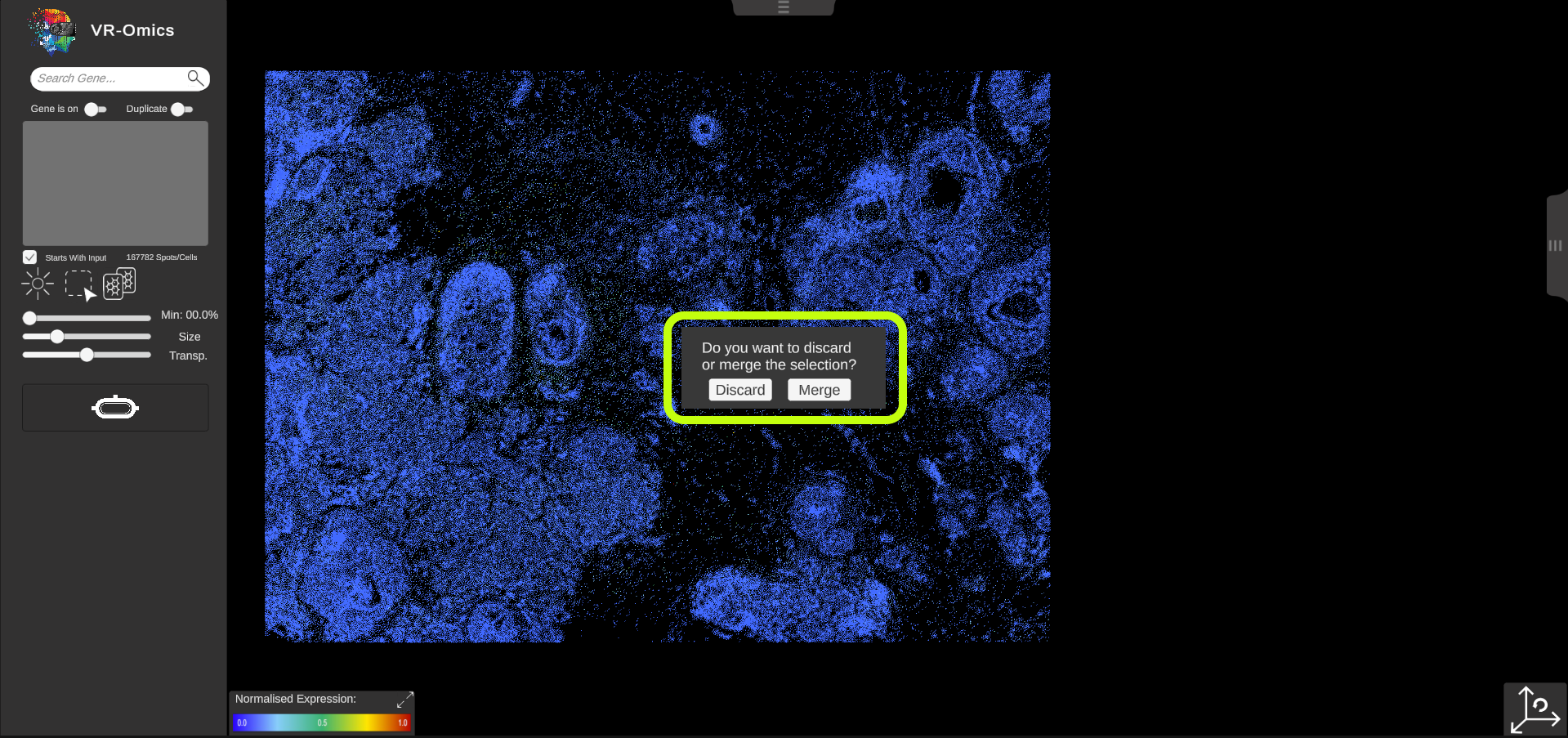
|
| Showing spatially variable genes (SVGs) results
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
If the SVG analysis was performed using the VR-Omics Autoamted Workflow, the Visualiser is able to highlight the SVGs in the search results. If a term is entered into the search bar, the autocomplete feature will show the search results for the available genes. The buttons for SVGs will be highlighted in cyan. |
|
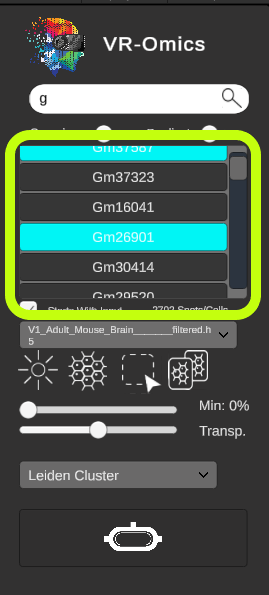
|
| Browse SVG results in the Visualiser
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
If the SVG analysis was performed using the VR-Omics Autoamted Workflow, the SVG results can be browsed from the Visualiser:
|
|
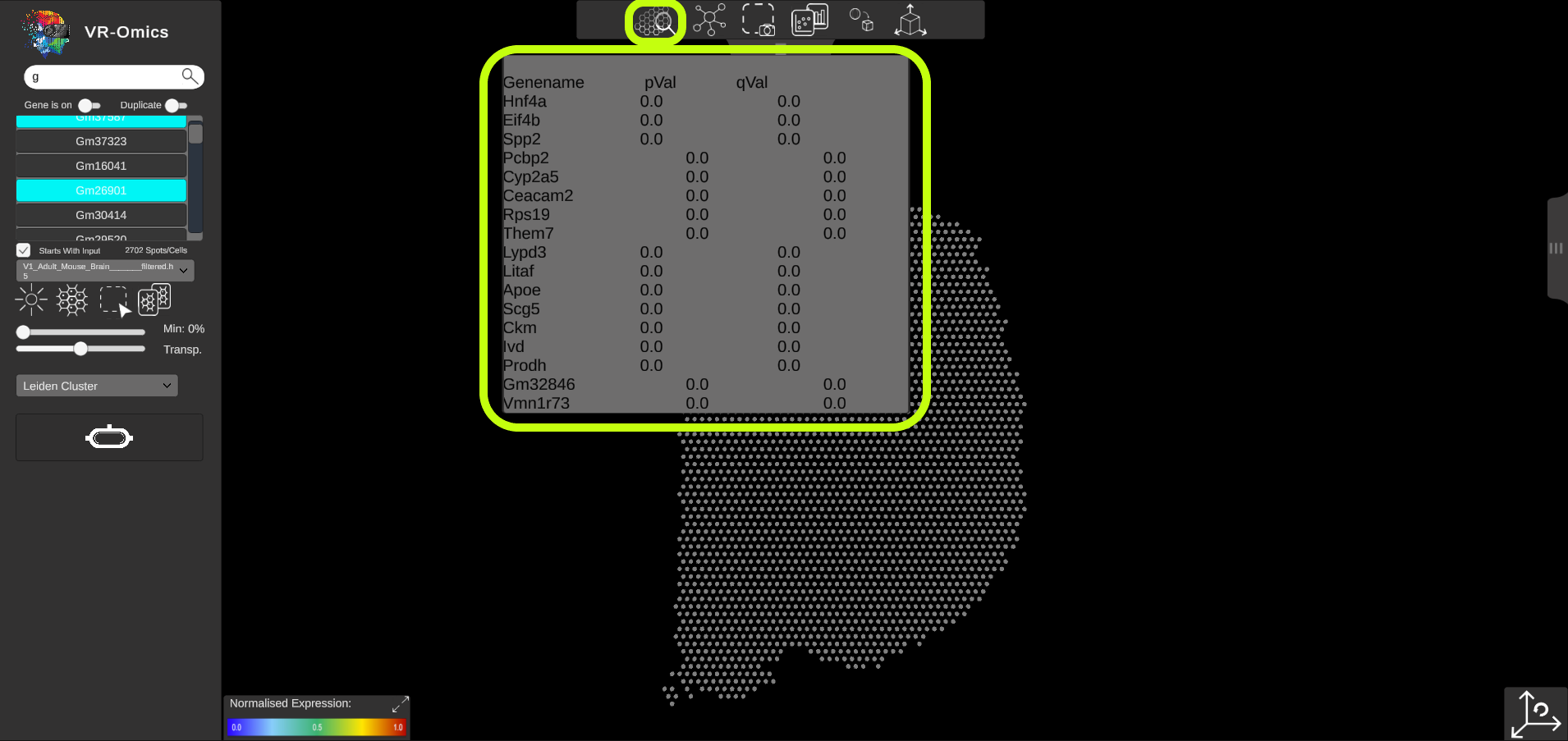
|
Export Features
| Feature | Description | Icon | Image |
|---|---|---|---|
| Continue Session
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
Your current session can be saved and continued to a later time point. This will include:
To save the session select the top menu and click on the To continue a saved session, from the main menu select the Keep in mind that saving the current session will override the last session saved. The sessions are saved as json files within the data structure. |
|
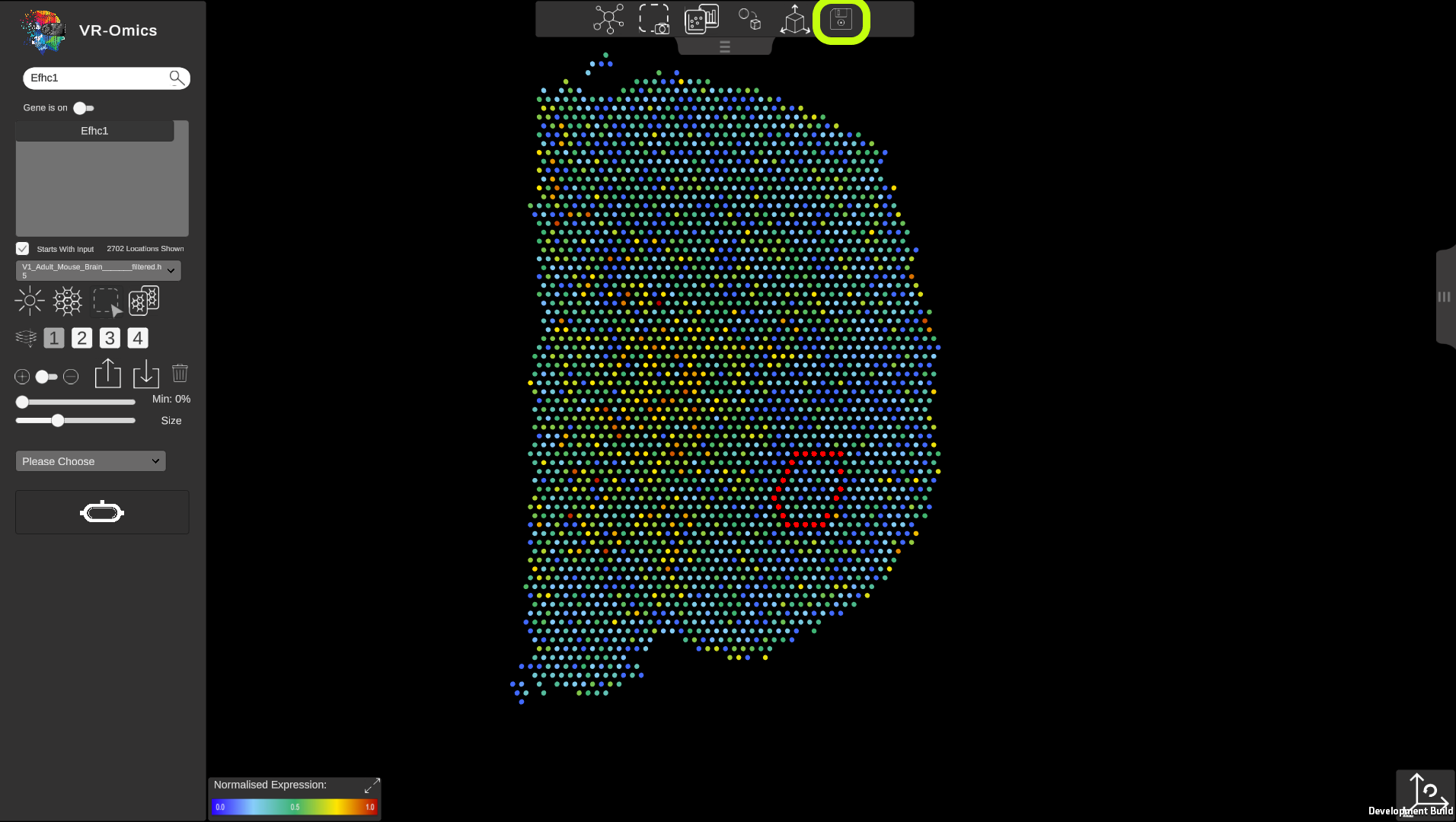
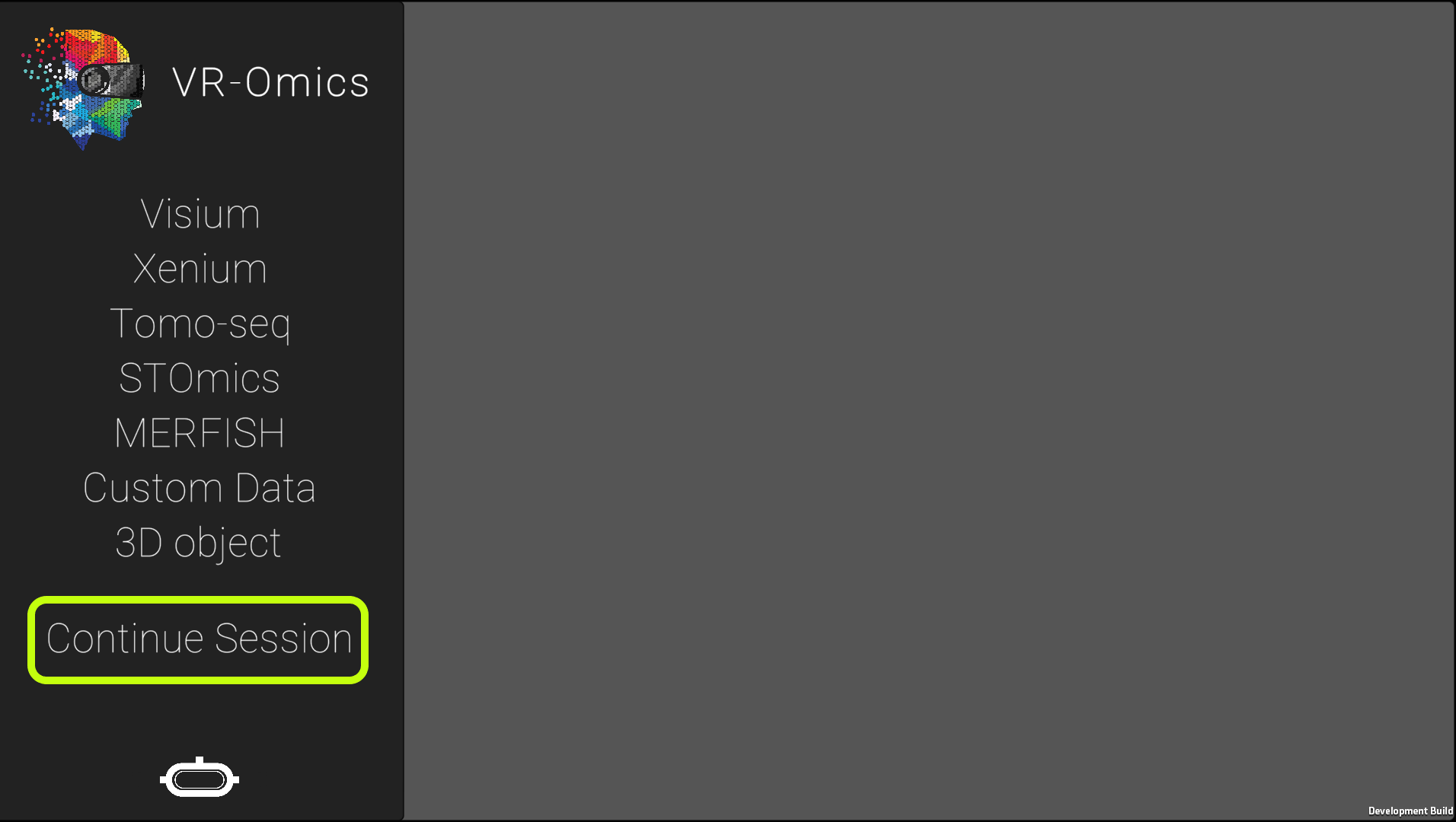
|
| Screenshot
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
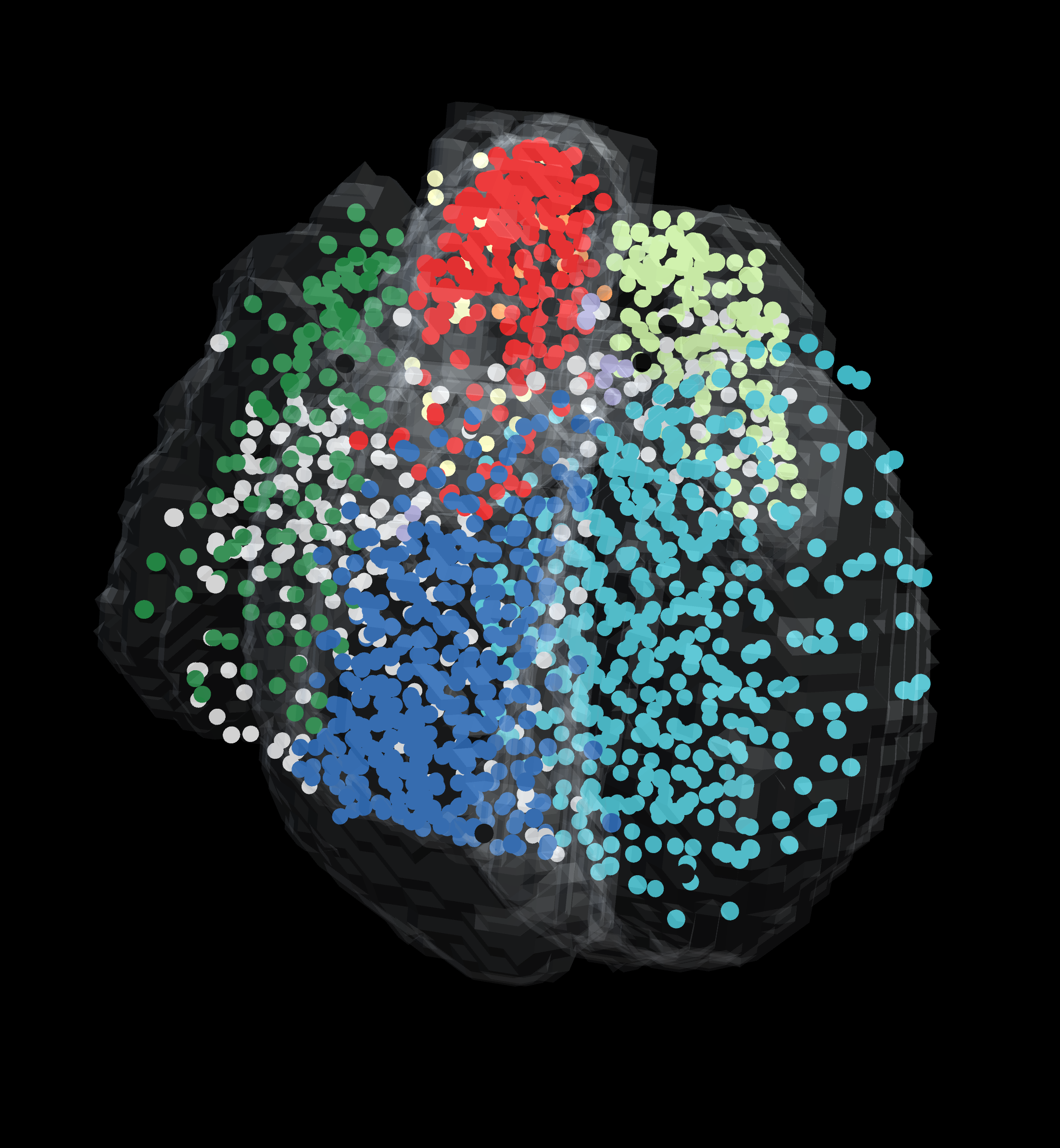
Screenshots can be made from the visualiser. For the screenshot only the dataset will be shown. To make a screenshot:
|
|
|
| Export ROI selections
Visium VR |
ROI selections can be exported. This will export all current selected locations and the respective groups they are in for later analysis. To export selections:
The export function will override the last selection. Save the selection locally by copying the export file.
|
|
|
| Import ROI selections
Visium VR |
To import previously saved ROI selections:
The export function does not check if the same dataset is used. Importing the last selection into a different dataset will cause errors. Ensure using the same data set to import as the one the export was generated from.
|
|
|
| Output plots
Visium VR |
If the VR-Omics Automated Workflow was used to download the Visium data optionally the output plots can be generated. The plots will be saved within the sample folder in the VR-Omics folder The plots can also be quick accessed from the top menu through the Visualiser |
|
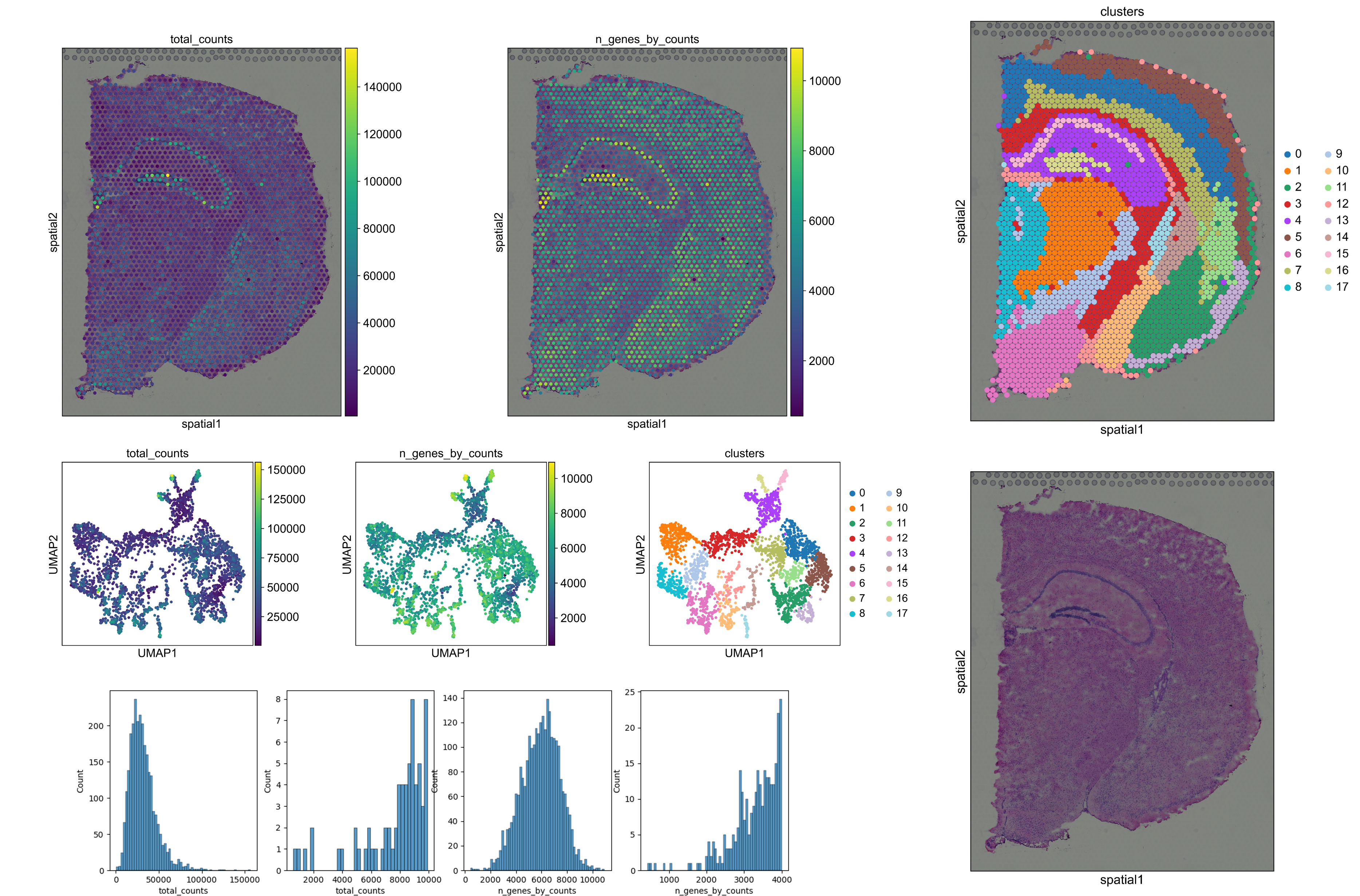
|
Switch VR and Desktop Application
| Feature | Description | Icon | Image |
|---|---|---|---|
| Switching between desktop and VR application
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
VR-Omics allows to easily switch between the desktop application and the VR environment:
We used the VR toolkit plugin (XR Interaction Toolkit version 2.0.3) supported are all devices that are supported by the toolkit. The following headsets have been confirmed to work with the plugin but not all are tested with VR-Omics: Windows Mixed Reality (eg: HP Reverb, Samsung Odyssey), Oculus Quest & Quest 2, Pimax 5K & 8KX, Varjo Aero & VR-3, Valve Index, HTC Vive, HTC Vive Cosmos, Pico Neo 3 & 4. We recommend using the Oculus Quest 2 or Quest Pro |
off on |

|
Additional Parameters to Explore
| Feature | Description | Icon | Image |
|---|---|---|---|
| Access spatial plots
Visium VR |
If spatial plots are available they can be visualised in the VR-Omics Visualiser: More information on which plots are available with the VR-Omics Automated Workflow can be found here To show the plots in the Visualiser:
|
|
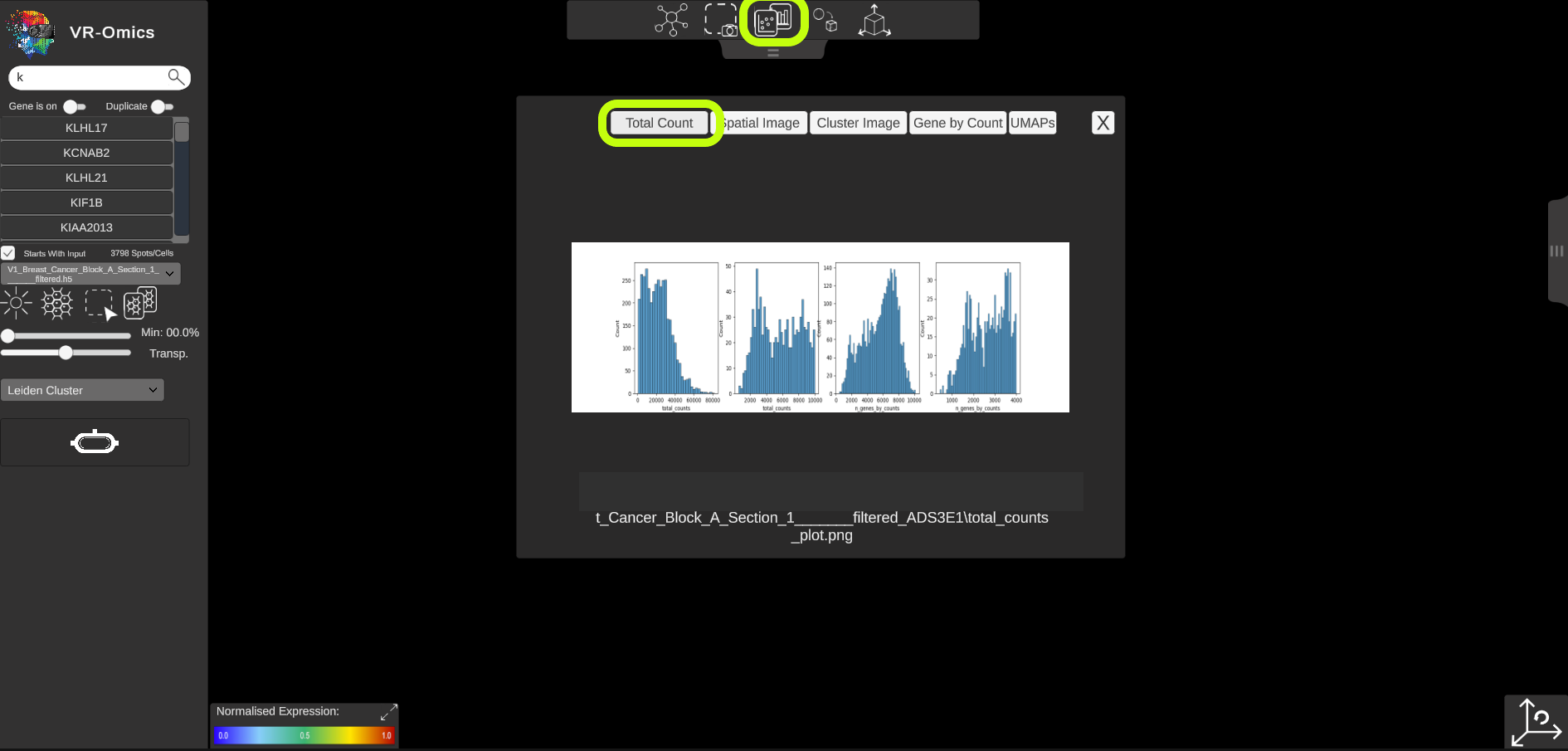
Show all available plots: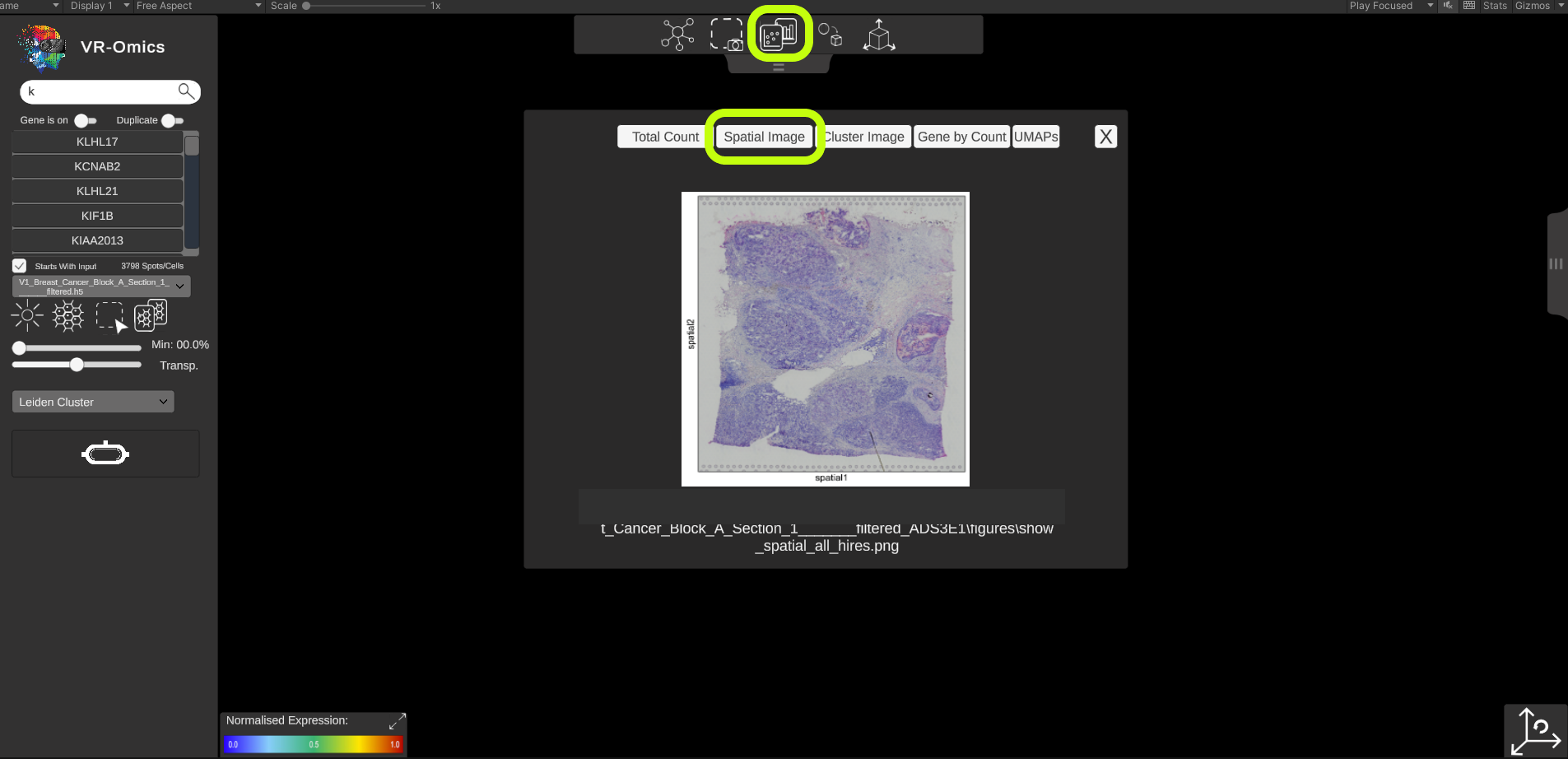
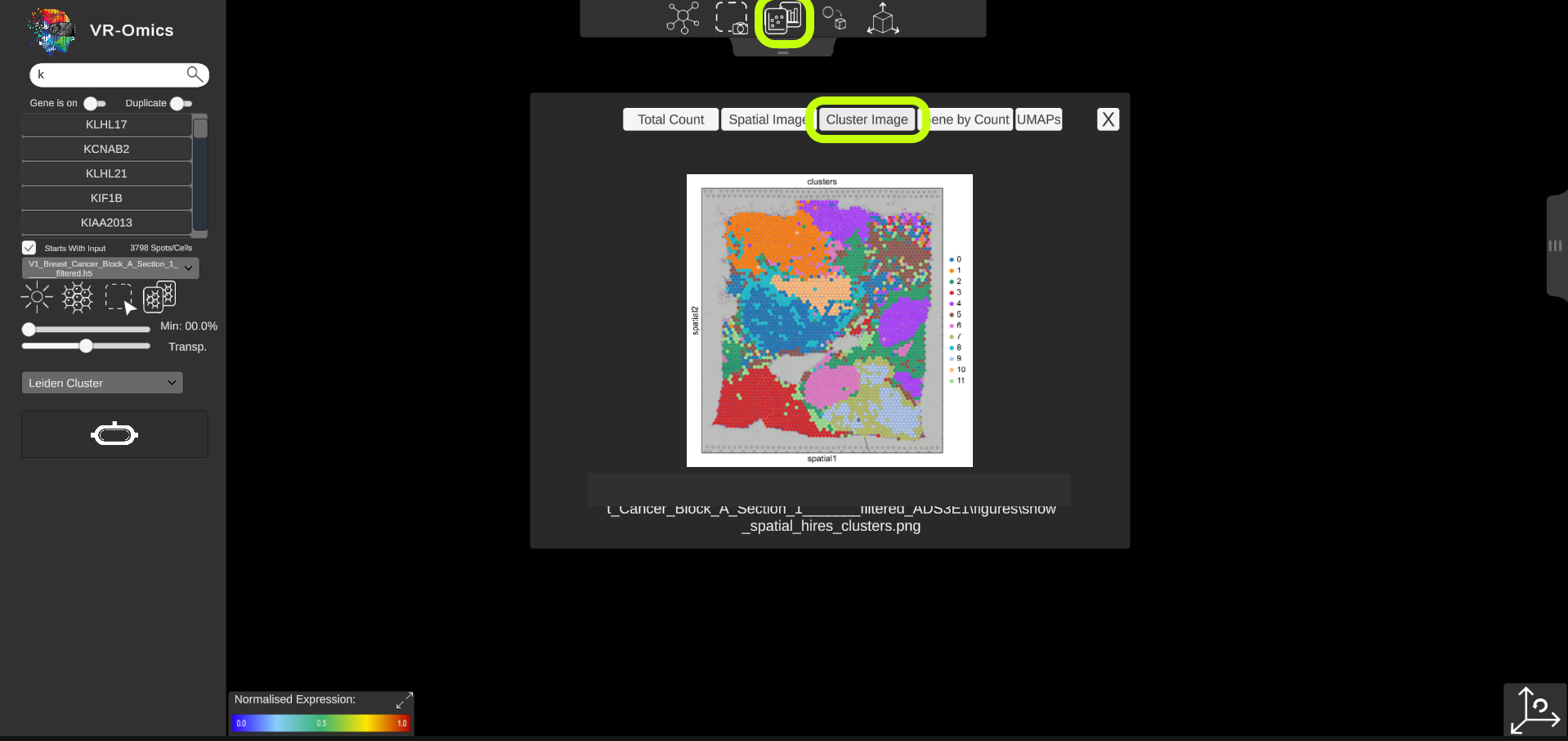
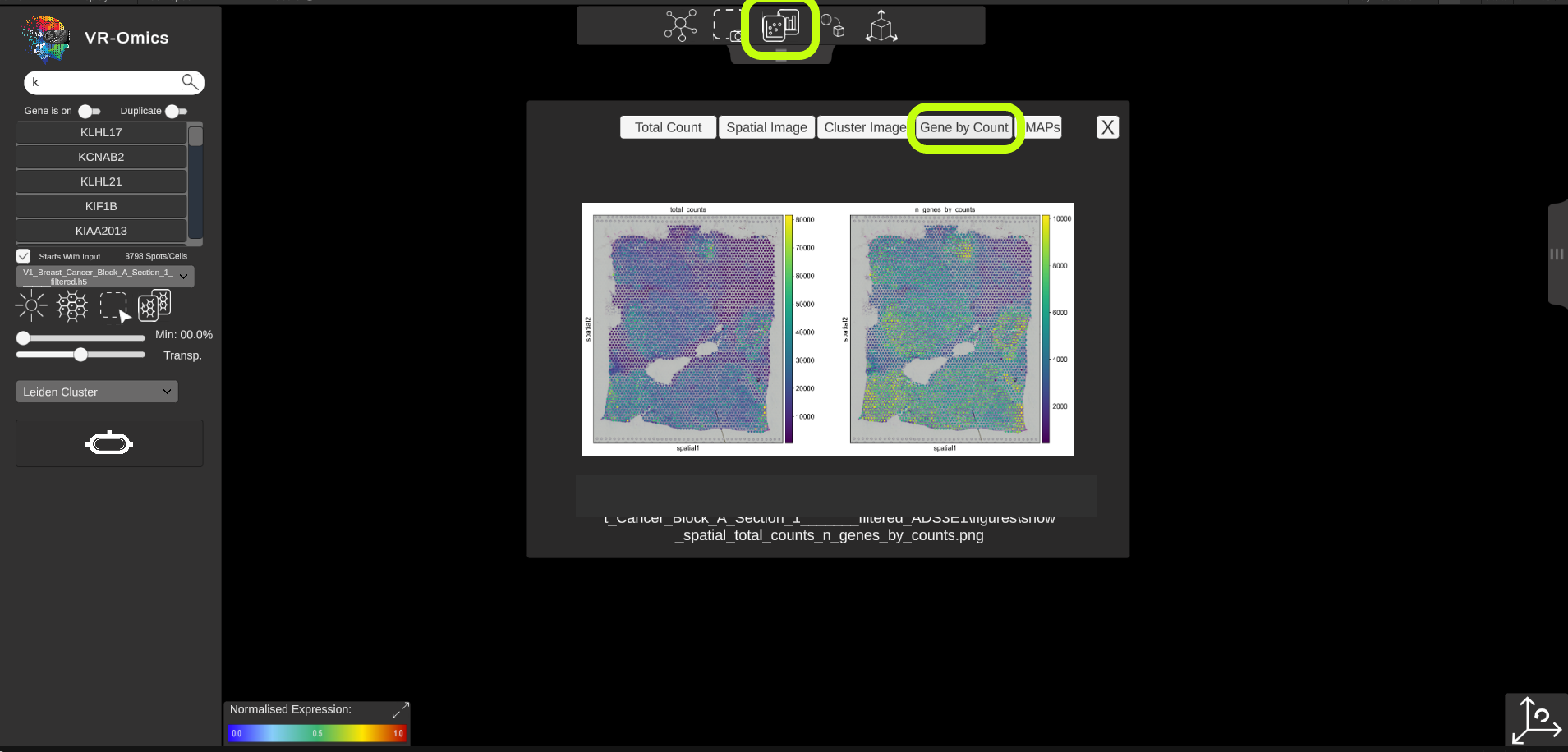
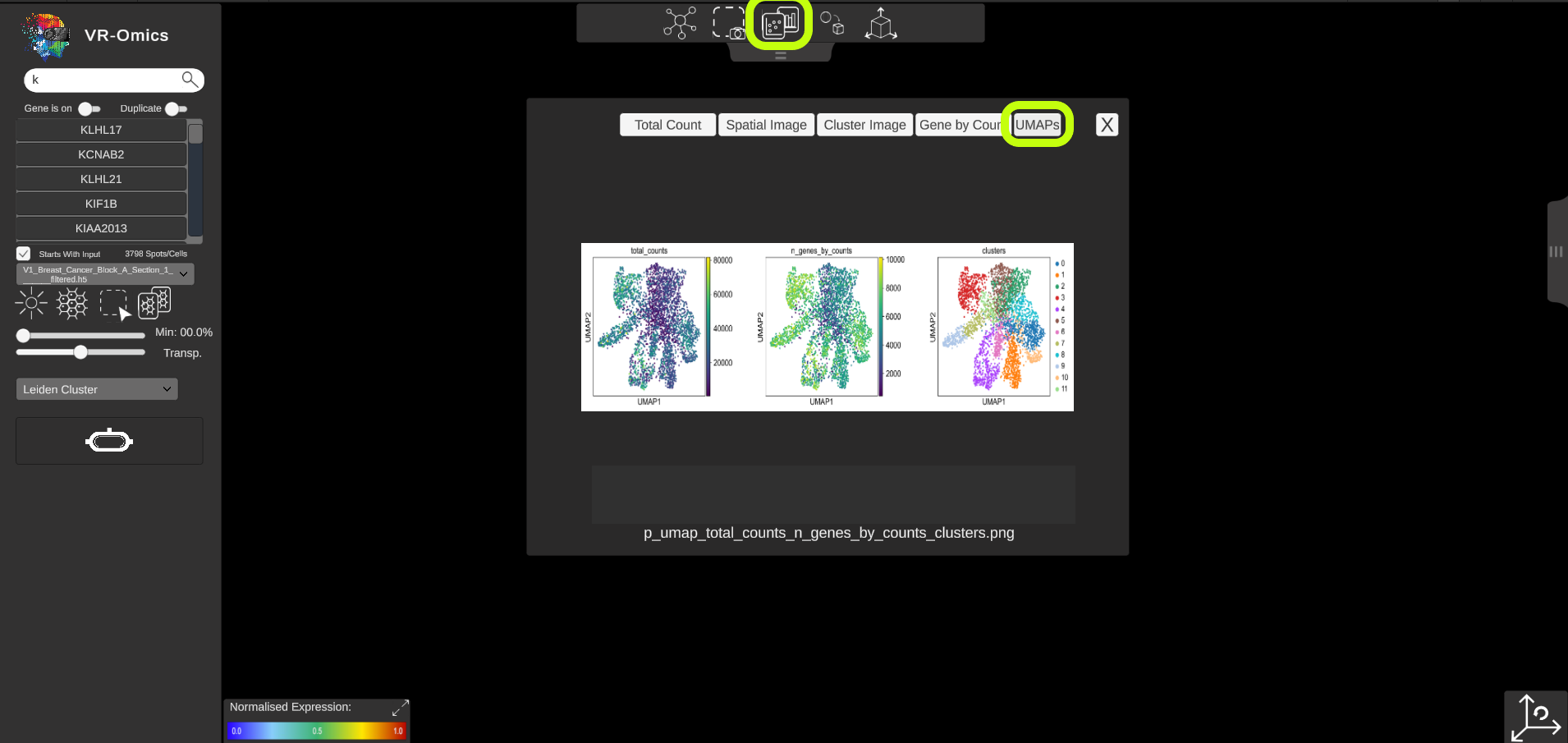
|
3D Object Alignment
| Feature | Description | Icon | Image |
|---|---|---|---|
| Change 3D Object Opacity
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
The opacity of the 3D object can be adjusted:
|
|


|
| Move 3D object
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
The 3D object can be moved independently from the dataset to align the object:
|
|

|
| Rotate 3D Object
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
The 3D object can be rotated independently from the dataset to align the object:
|
|
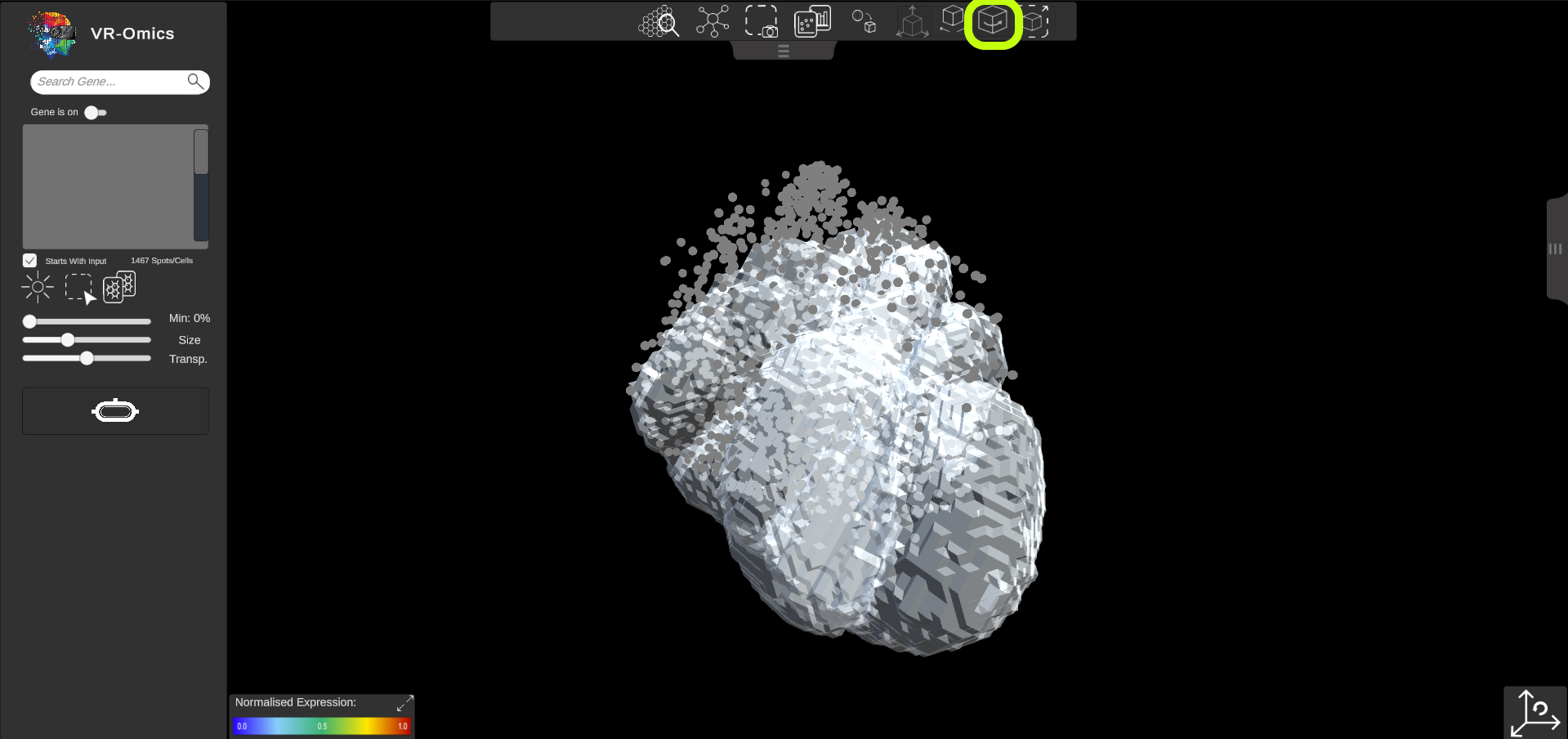
|
| Resize 3D Object
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
The 3D object can be resized independently from the dataset to align the object:
|
|
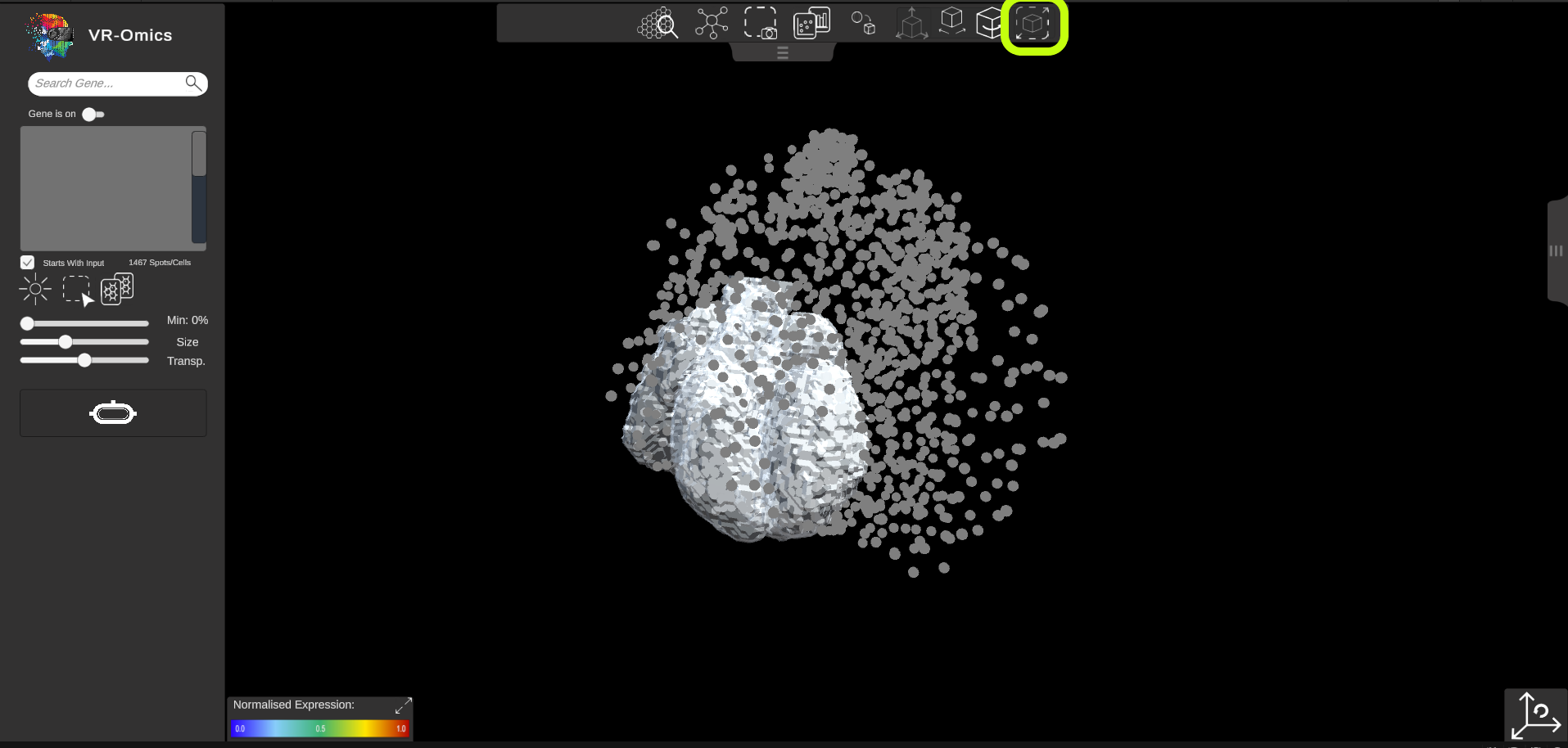
|
Customisation
| Feature | Description | Icon | Image |
|---|---|---|---|
| Change the Symbol
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
Per default the Symbol to visualise a location is a sphere (except of Tomo-Seq). This can be changed to a Cube:
|
|

|
| Change the Symbol Size
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
The symbol size can be changed:
This feature is disabled for some technologies like Visium since resizing the spots would distort the visualisation of the data.
|
Size
|
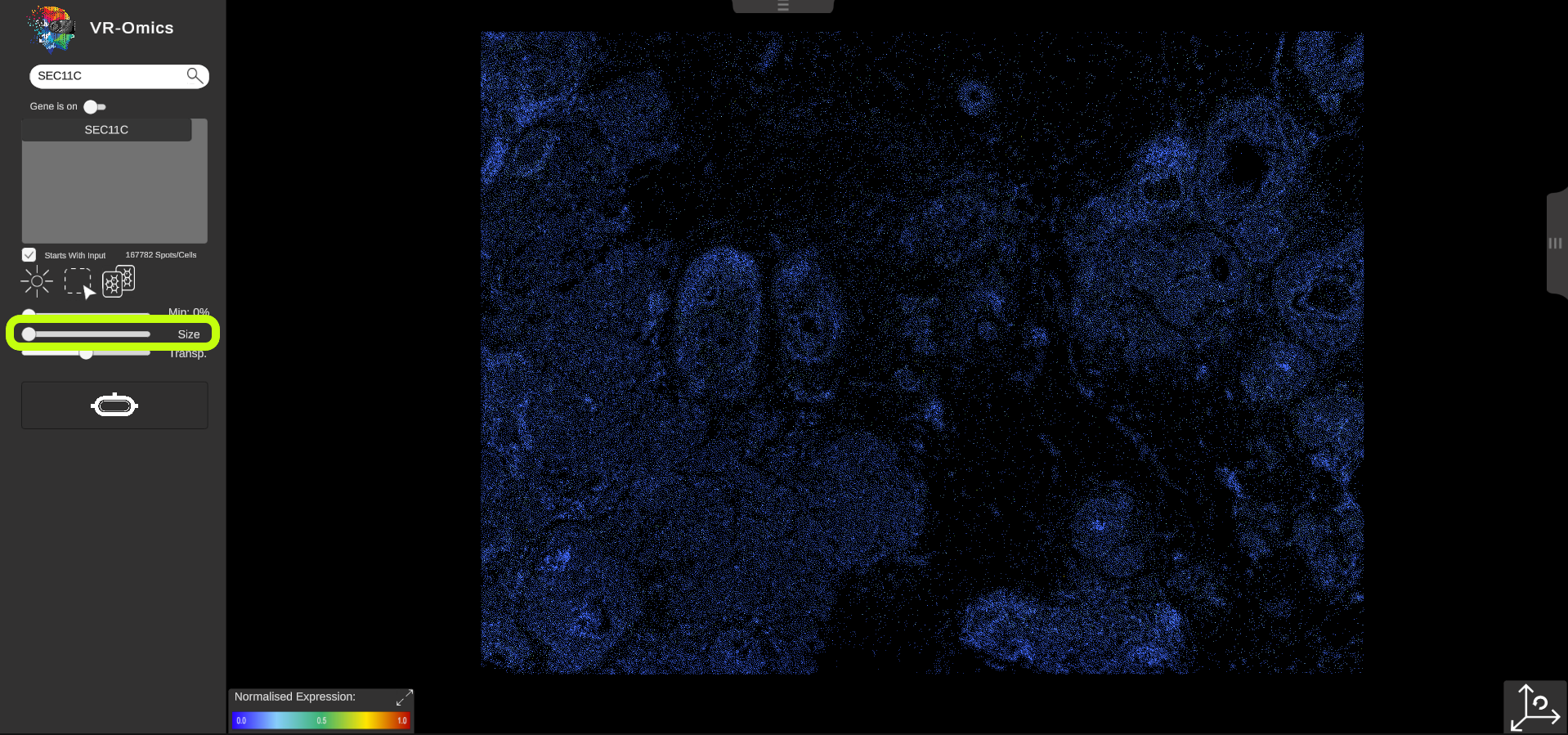
|
| Reset the view
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
To reset the view to the origin click the reset view button in the bottom left of the screen. |
|
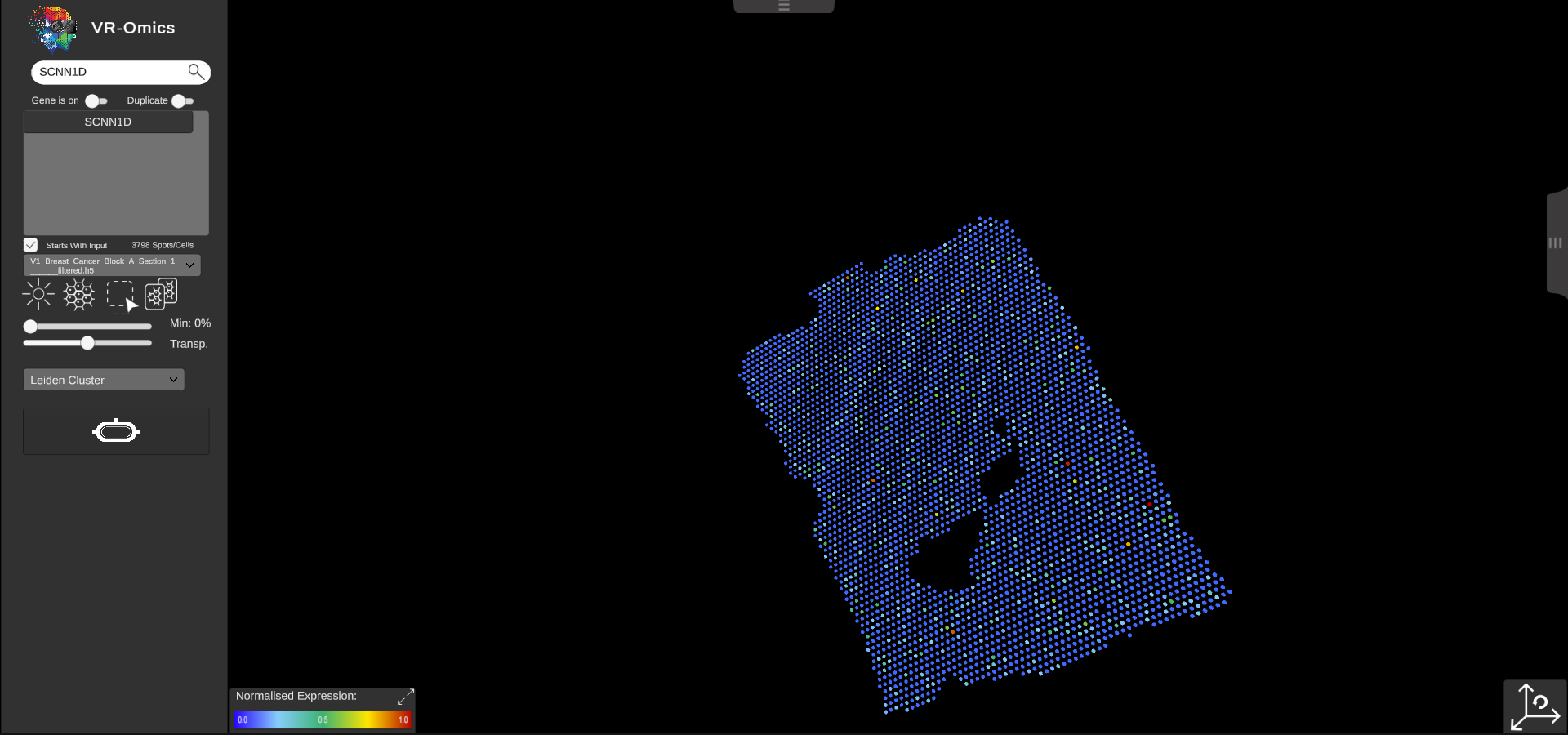
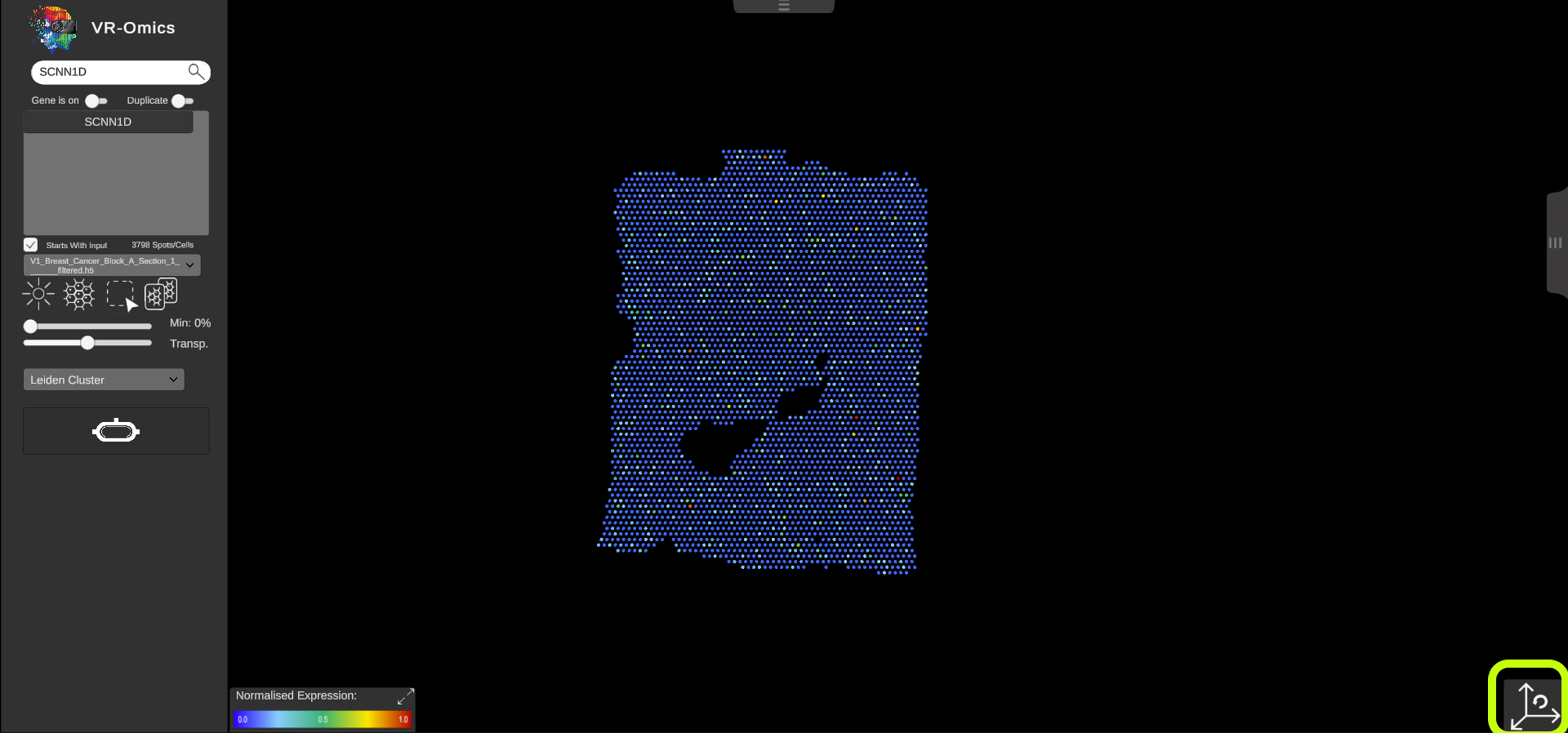
|
| Change Background Colour
Visium Xenium STOmics Merfish Tomo-Seq Custom VR Demo |
The background colour can be changed from dark to bright mode.
|
|
|
Address
Murdoch Children's Research Institute, Parkville, Melbourne
denis.bienroth@mcri.edu.au