Explore our Software
VR-Omics
Software to analyse and explore spatially resolved transcriptomics data. Available as desktop solution or Virtual environment.
VR-Cardiomics
Virtual Reality environment to explore spatial gene expression of mouse heart data. Supporting VR-headsets.
VR-Cardiomics for zSpace
Virtual Reality environment to explore spatial gene expression of mouse heart data. Supporting a zSpace device
3DCardiomics
Explore spatial gene expression of the mouse heart on your local machine. Visit the 3DCardiomics web application.
MonaGo
MonaGO is a visualization tool for Gene Ontology (GO) enrichment analysis. Visit the MonaGo web application.
Enhancer Workflow
EnhancerWorkflow - A workflow to screen enhancers using cis-regulatory elements (CREs)
Trawler-2.0
Discover over-represented motifs in chromatin immunoprecipitation (ChIP) experiments and to predict their functional instances.
cNRE-Genomics-Analysis
Comparative genomics analyses of cNRE (complex nuclear receptor cis-regulatory element)
Cardiac Network Component Predictor
A pipeline for predicting cardiac gene network components using cis-regulatory elements (CREs)
About Us
Transcriptomics and Bioinformatics
Our vision is to data-mine our genetics information to unravel the developmental gene regulatory networks that are required to form healthy babies, and that cause congenital malformations if altered.
The Transcriptomics & Bioinformatics group are a multidisciplinary team of computational and molecular biologists who specialised in mining our genomic information, in order to uncover the underlying genetic causes of congenital diseases, such as congenital heart disease (CHD). The group is particularly focused on decrypting the role of the non-coding genome in development and disease, via developing novel software, data mining pipelines and conducting bioinformatics research in collaboration with other laboratories at MCRI and beyond.
Challenges facing children and adolescents
The successful formation of healthy babies relies on a network of genes that must be activated at exact times and in specific territories during embryonic development. Failure to activate this network at the right time and right place will result in congenital malformations. Therefore, it is imperative that we 1) identify the genetic components of this gene regulatory network, and 2) understand where and when these genes are recruited, in order to fully understand how healthy babies develop and how genetic malformations may arise.
Our current research
To address this challenge, the research team applies systems biology approaches (the study of biological components, be it molecules, cells, organisms or entire species), to reconstruct the developmental gene regulatory networks that are required to form healthy babies, and that cause congenital malformations if altered. The group develops novel software for -omics data mining (genomics end epigenomics, single cell and spatial transcriptomics, networks) and 3D data visualisation platforms, making complex high-throughput data intuitive and interpretable by expert researchers, clinicians and lay audiences alike. Most importantly, the group collaborates with life scientists all around the world to exploit the power of Bioinformatics to enable breakthrough discoveries in our understanding of embryonic development and congenital disease.
Future impact on children and adolescents
Our bioinformatics research delivers enabling bioinformatics resources to collaborating laboratories, who employ these resources to drive better insights into genes and their contributions to health and diseases in babies. Our data mining research also helps derive gene panels with proven predictive power which are essential for producing reliable diagnostics tests for parents and babies.
Our Latest Publications
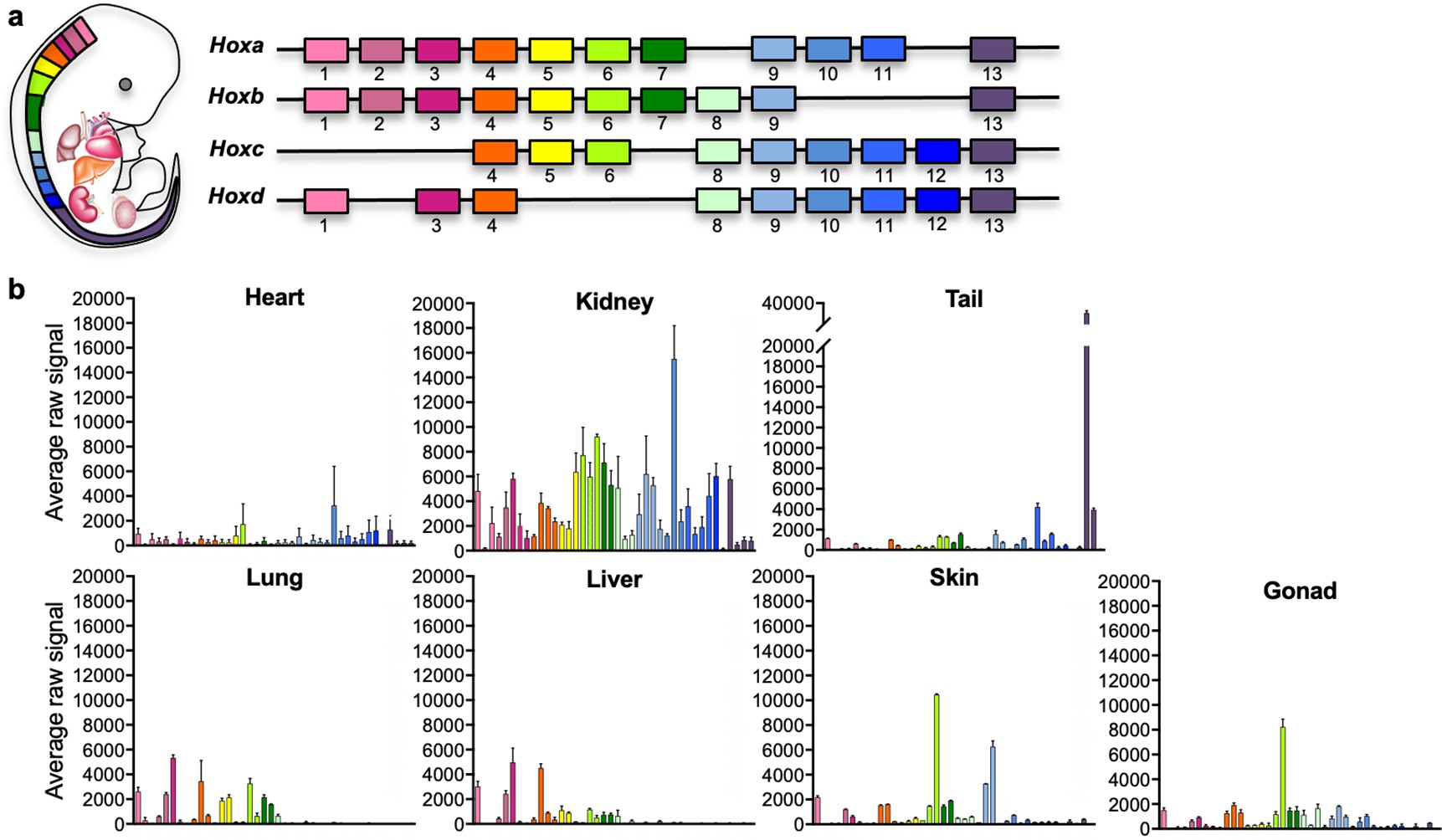
Adult mouse fibroblasts retain organ-specific transcriptomic identity
Organ fibroblasts are essential components of homeostatic and diseased tissues. They participate in sculpting the extracellular matrix, sensing the microenvironment, and communicating with other resident cells. Recent studies have revealed transcriptomic heterogeneity among fibroblasts within and between organs. To dissect the basis of interorgan heterogeneity, we compare the gene expression of murine fibroblasts from different tissues (tail, skin, lung, liver, heart, kidney, and gonads) and show that they display distinct positional and organ-specific transcriptome signatures that reflect their embryonic origins. We demonstrate that expression of genes typically attributed to the surrounding parenchyma by fibroblasts is established in embryonic development and largely maintained in culture, bioengineered tissues and ectopic transplants. Targeted knockdown of key organ-specific transcription factors affects fibroblast functions, in particular genes involved in the modulation of fibrosis and inflammation. In conclusion, our data reveal that adult fibroblasts maintain an embryonic gene expression signature inherited from their organ of origin, thereby increasing our understanding of adult fibroblast heterogeneity. The knowledge of this tissue-specific gene signature may assist in targeting fibrotic diseases in a more precise, organ-specific manner.
Detection and identification of cis-regulatory elements using change-point and classification algorithms
FTranscriptional regulation is primarily mediated by the binding of factors to non-coding regions in DNA. Identification of these binding regions enhances understanding of tissue formation and potentially facilitates the development of gene therapies. However, successful identification of binding regions is made difficult by the lack of a universal biological code for their characterisation.

Spatially resolved transcriptomics in immersive environments
Spatially resolved transcriptomics is an emerging class of high-throughput technologies that enable biologists to systematically investigate the expression of genes along with spatial information. Upon data acquisition, one major hurdle is the subsequent interpretation and visualization of the datasets acquired. To address this challenge, VR-Cardiomics is presented, which is a novel data visualization system with interactive functionalities designed to help biologists interpret spatially resolved transcriptomic datasets. By implementing the system in two separate immersive environments, fish tank virtual reality (FTVR) and head-mounted display virtual reality (HMD-VR), biologists can interact with the data in novel ways not previously possible, such as visually exploring the gene expression patterns of an organ, and comparing genes based on their 3D expression profiles. Further, a biologist-driven use-case is presented, in which immersive environments facilitate biologists to explore and compare the heart expression profiles of different genes.
Address
Murdoch Children's Research Institute, Parkville, Melbourne
denis.bienroth@mcri.edu.au